Membrane Proteins - Indispensable Gatekeepers
Membrane proteins are proteins associated with or attached to the cellular membrane of cell compartments or organelles of a cell. They represent one of the largest and most important classes of proteins and can be classified as either peripheral or integral.
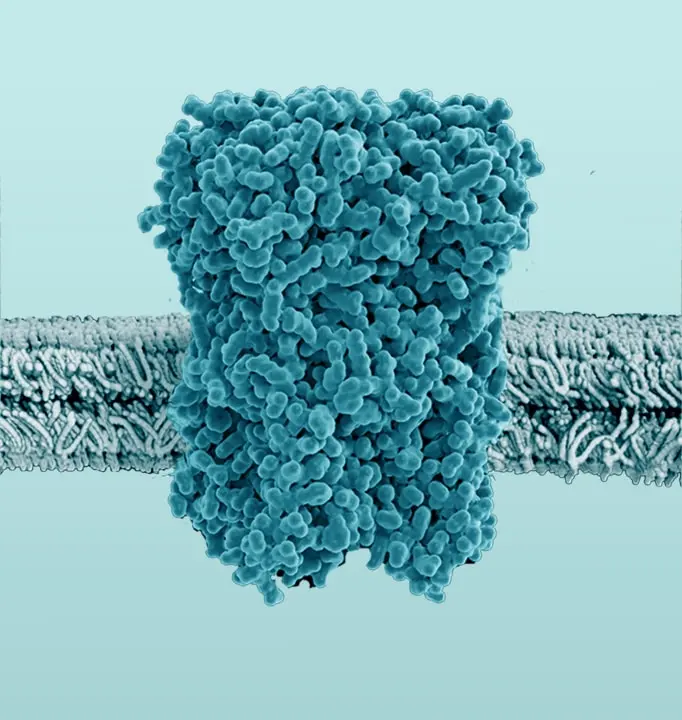
The Membrane Proteome
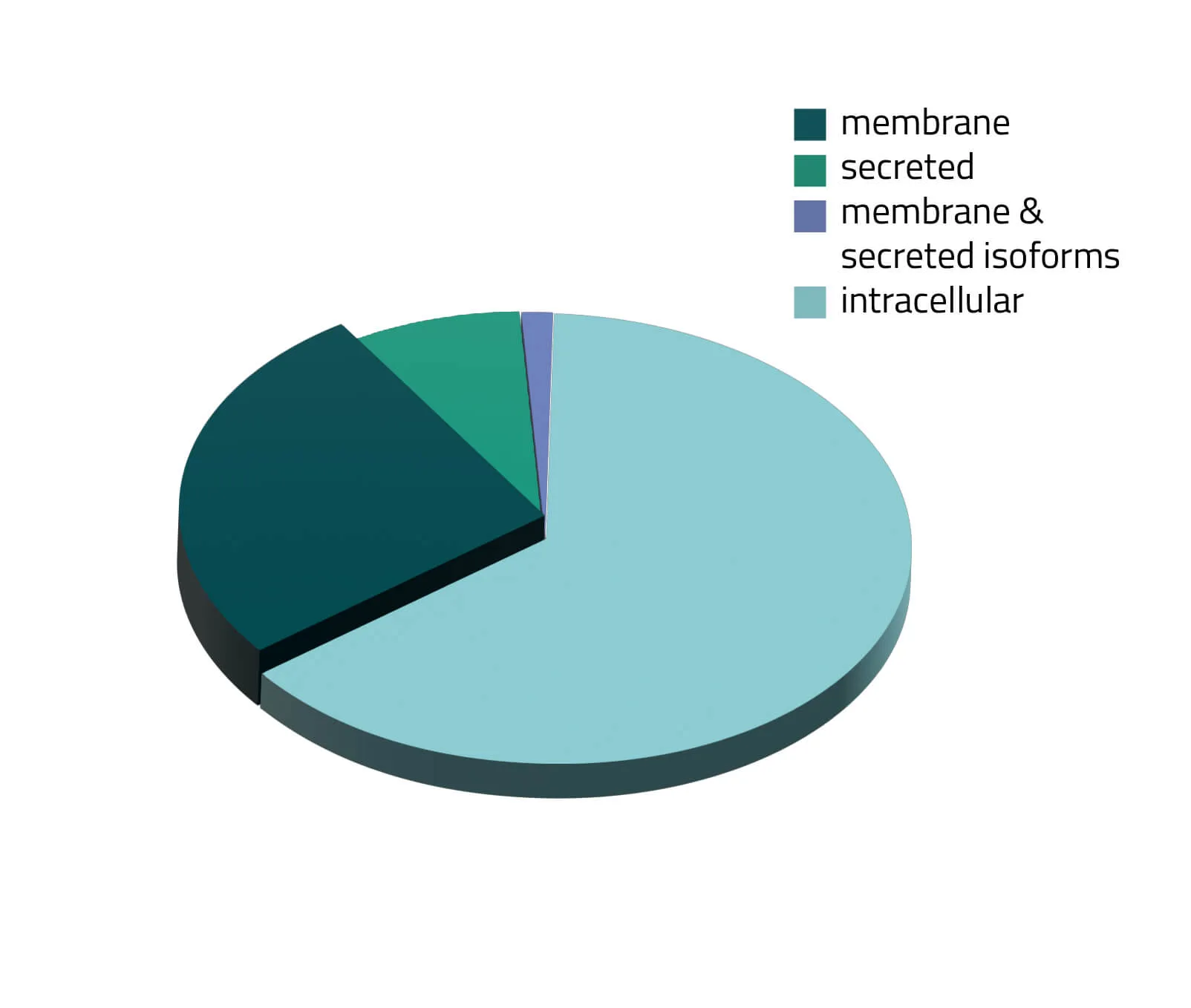
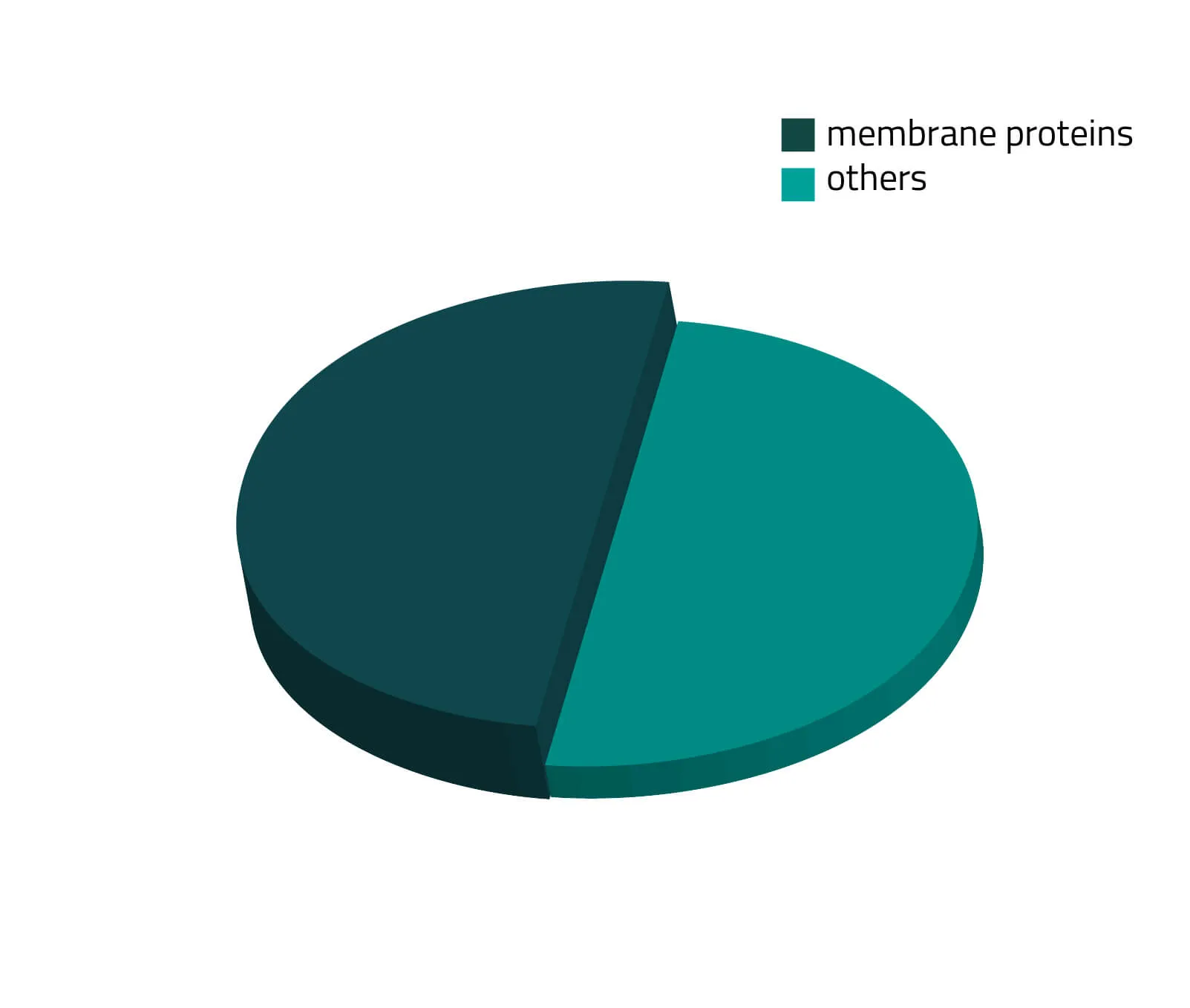
Over the past decades, the number of known human protein-coding genes has always shifted slightly, but in recent years the absolute number is revolving around 20.000 genes (Piovesan, Antonaros and Vitale et al. 2019; Salzberg, 2018). Around one third of those proteins are secreted or membrane-bound proteins (see Figure 1). Although this being a significant portion of the whole proteome, only a few of them are structurally known. As about half of all approved therapeutics target membrane proteins (see Figure 2), resolving the structure of these therapeutically relevant membrane proteins is highly favorable for future drug design (Baker, 2010).
What is a Membrane Protein?
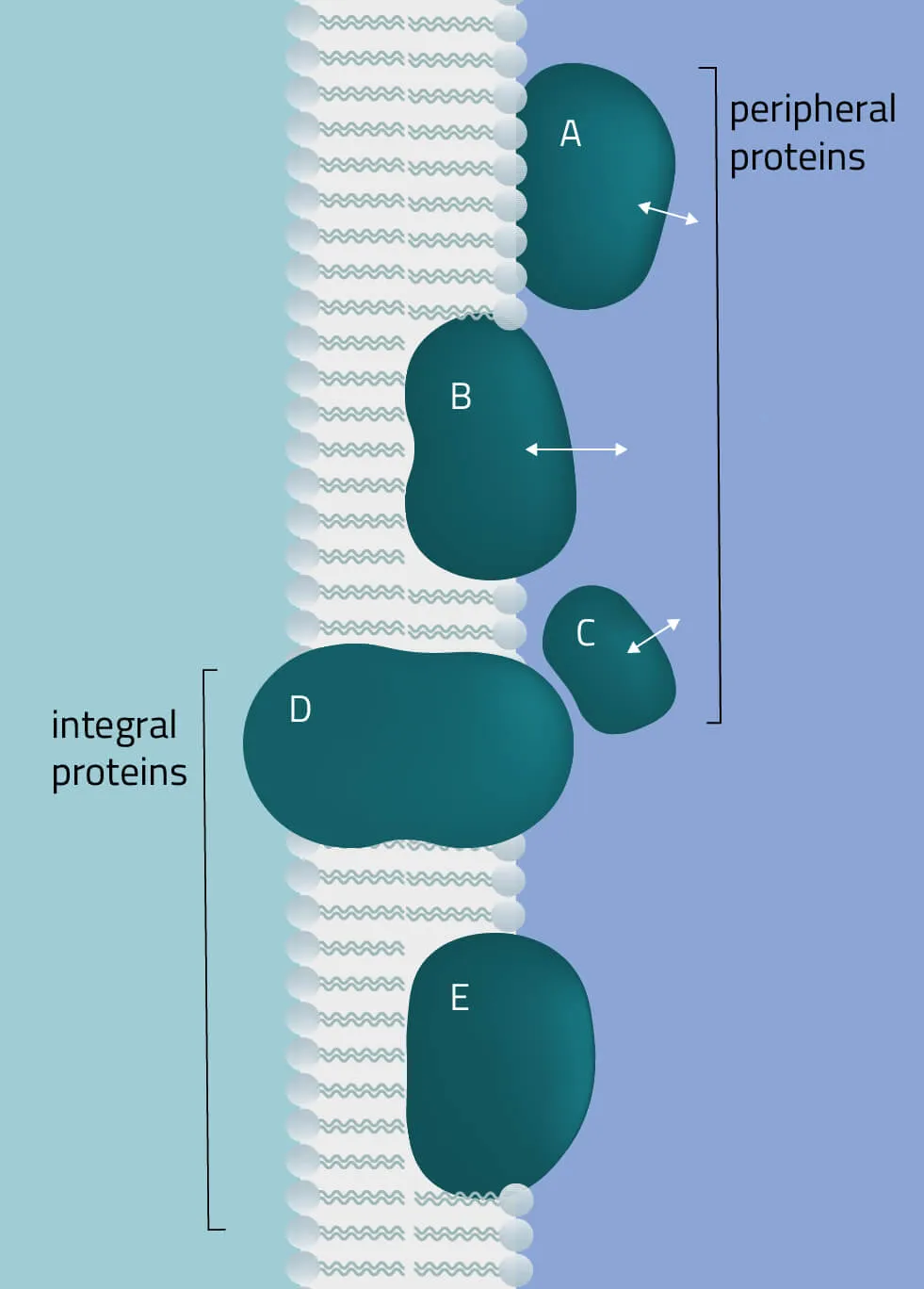
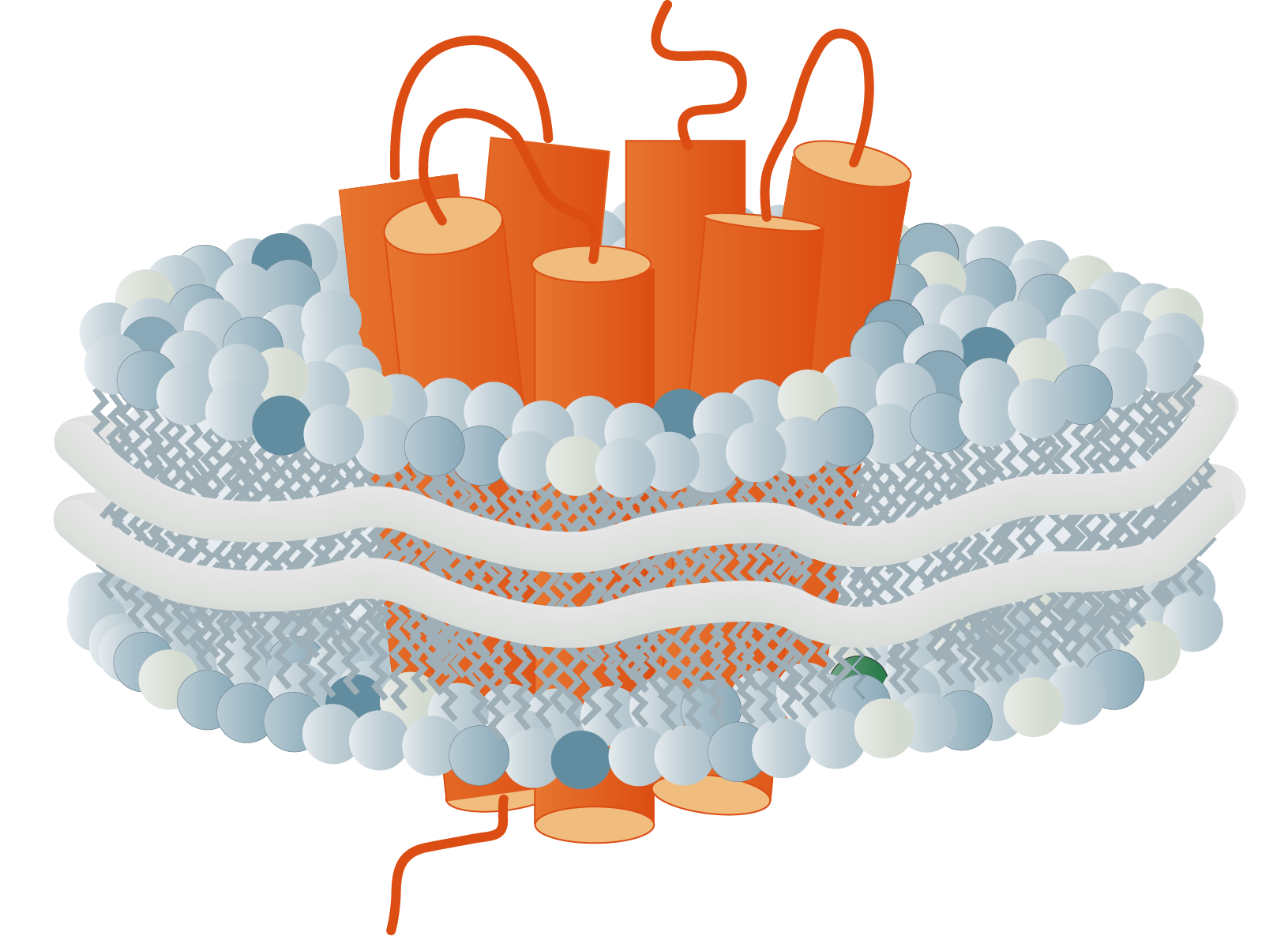
The membrane proteome and secretome are counted as one of the largest and most important classes of proteins. Membrane proteins are defined as proteins being associated or attached to the membrane of a cell or an organelle inside the cell. They are divided into peripheral and integral proteins. Peripheral membrane proteins are temporally associated with the lipid bilayer but do not fully span the membrane. Attachment to the lipid bilayer is achieved by perforation of peripheral regions or by coupling with integral membrane proteins (see Figure 3, B & C). These integral proteins are permanently embedded, span the entire lipid bilayer, and contain hydrophobic alpha-helical or beta-barrel structures residing inside the membrane. They can be further subdivided into groups such as receptors or channels depending on their cellular function.
Together with hydrophilic extra- and intracellular domains, the majority of membrane proteins show an amphipathic character. The amphipathic character additionally produces a signature by which integral membrane proteins can often be identified. This is due to their primary structures containing 19-23 hydrophobic amino acids in their linear sequences, required to span the hydrophobic interior of a membrane. β-barrels with hydrophobic residues pointing to the outside of the barrel can also act as a good indicator of a membrane protein (Yeagle, 2016, The Membranes of Cells).
The critical role of membrane proteins
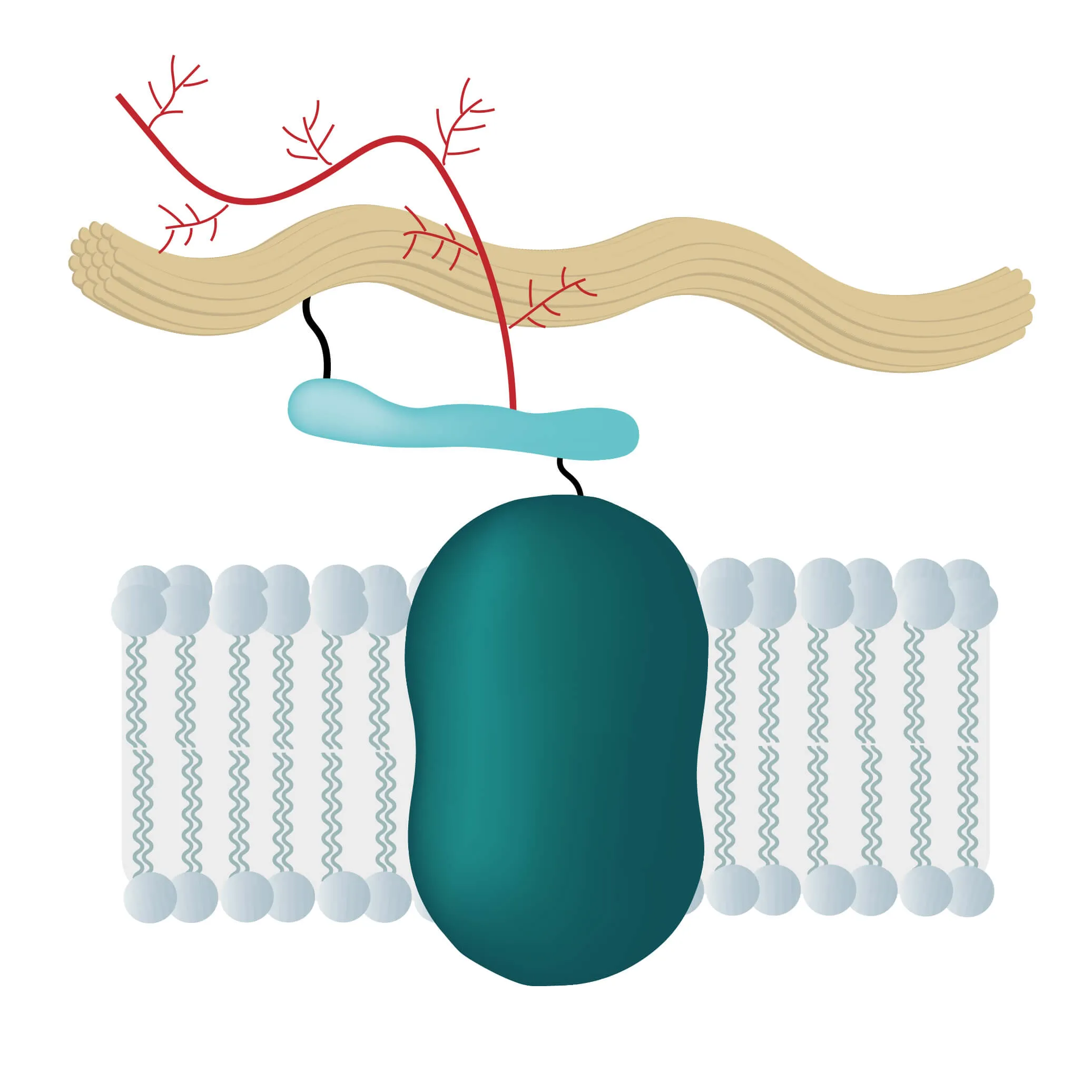
In many physiological and pathological processes reagents, transcription factors, proteins or ions need to pass membrane barriers. If successful they trigger signal pathways, send growth and coagulation factors or pass on signals of proteins not able to cross the lipid bilayer, like cytokines. Therefore, membrane proteins are located at a very important intersection of many cellular processes.
To do this, membrane proteins take over numerous critical functions like the transport of proteins and ions through special channels or the signal transduction inside many organisms. They are responsible for intercellular junctions or cell-cell recognition as well, enabling swift communication between cells and the effective recognition of foreign cells, being of utmost importance for the immune system.
To do this, membrane proteins take over numerous critical functions like the transport of proteins and ions through special channels or the signal transduction inside many organisms. They are responsible for intercellular junctions or cell-cell recognition as well, enabling swift communication between cells and the effective recognition of foreign cells, being of utmost importance for the immune system.
Classification and Functions of Membrane Proteins
Classes
Currently, the TCDB (Transporter Classification Database) lists 92 superfamilies with more than 1600 transporter families in them. Due to the sheer number of this, we would only like to present a small overview of these transporter classes in the following:
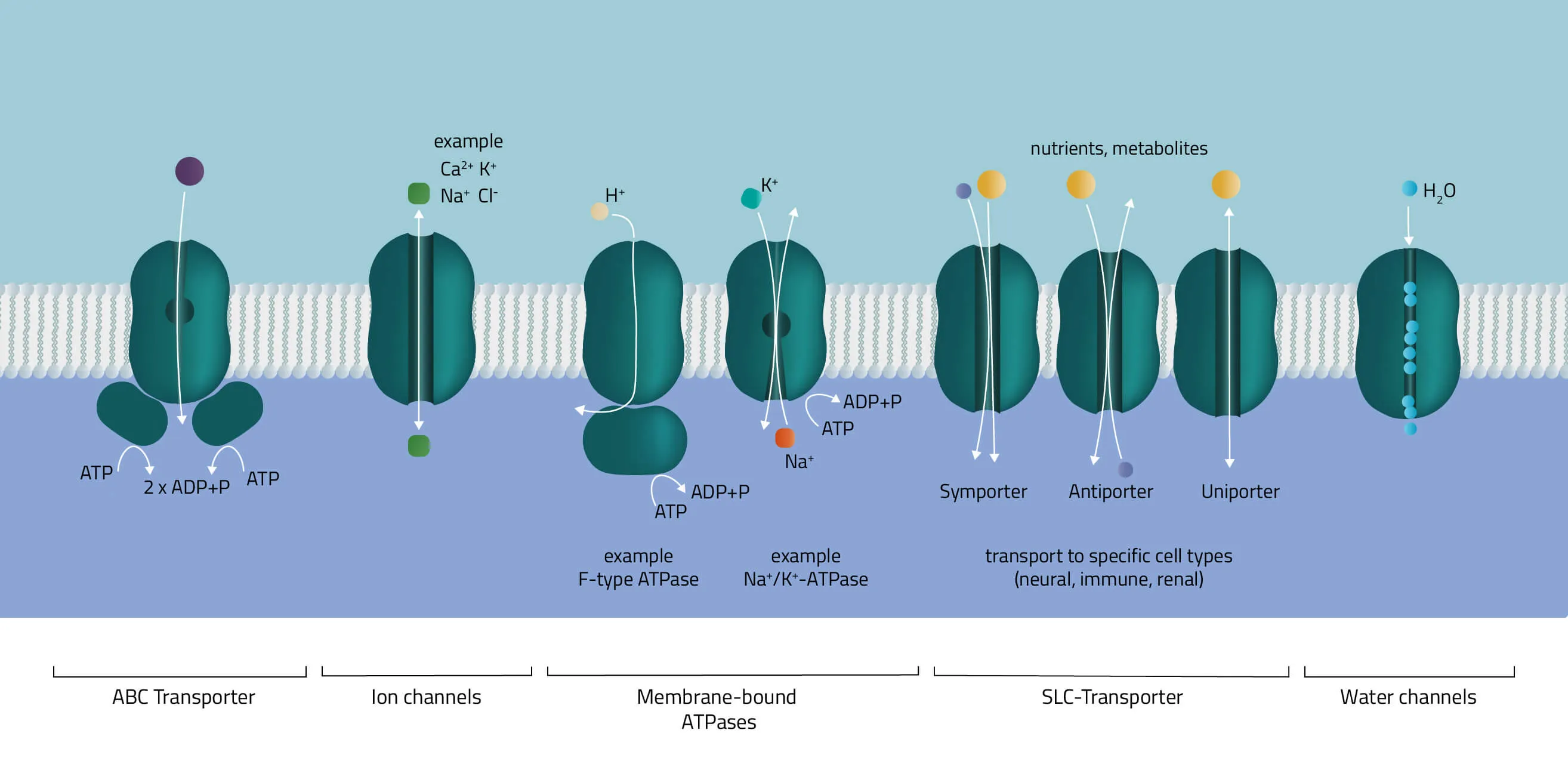
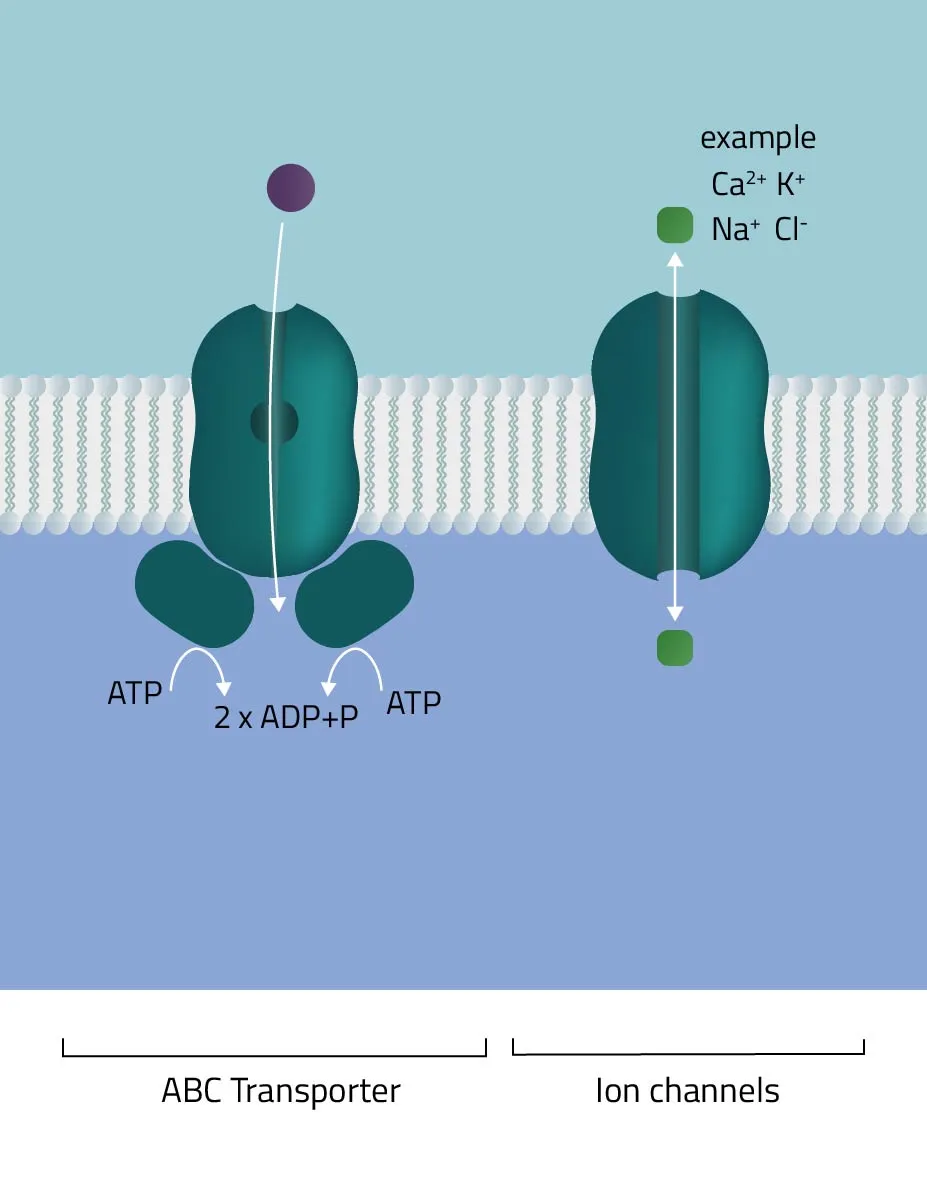
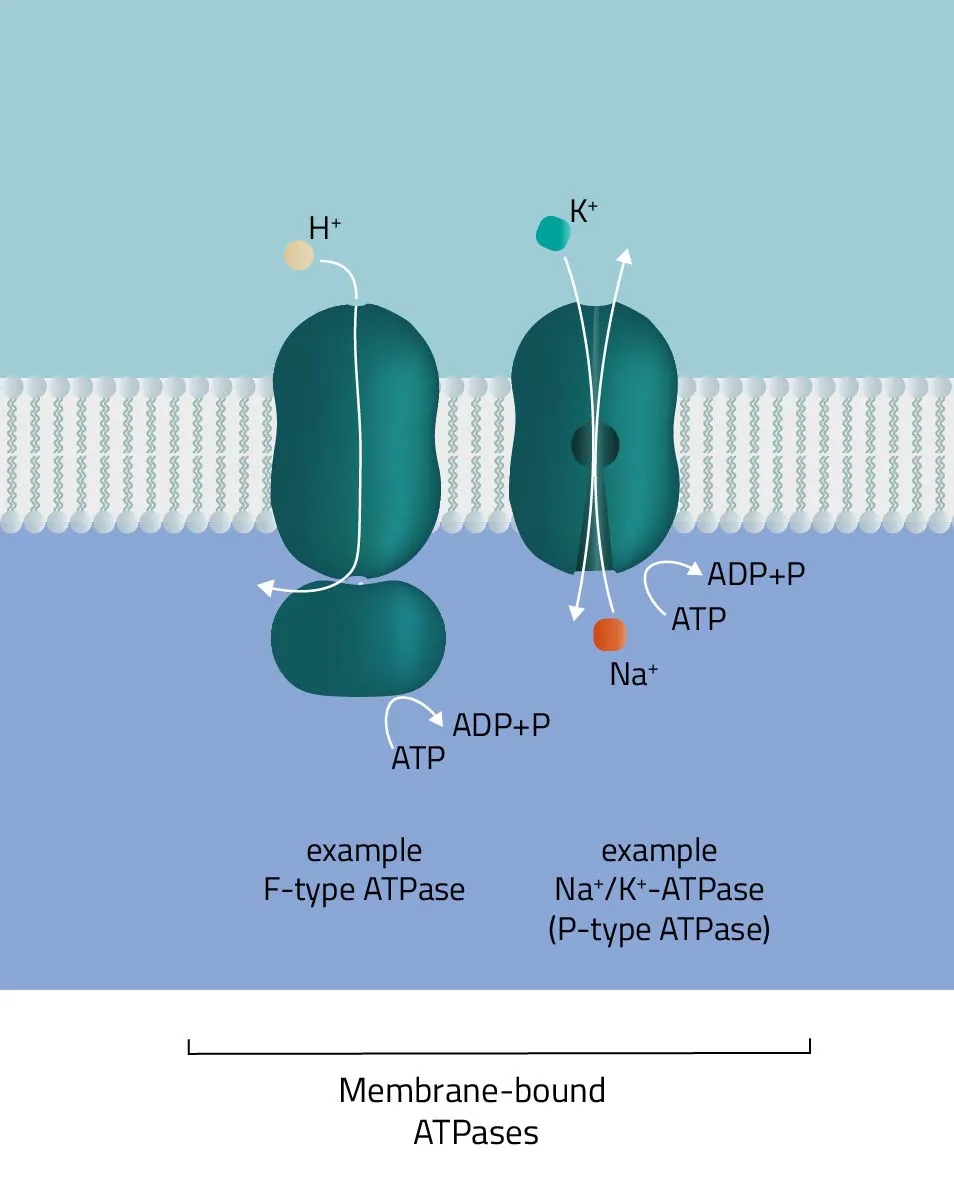
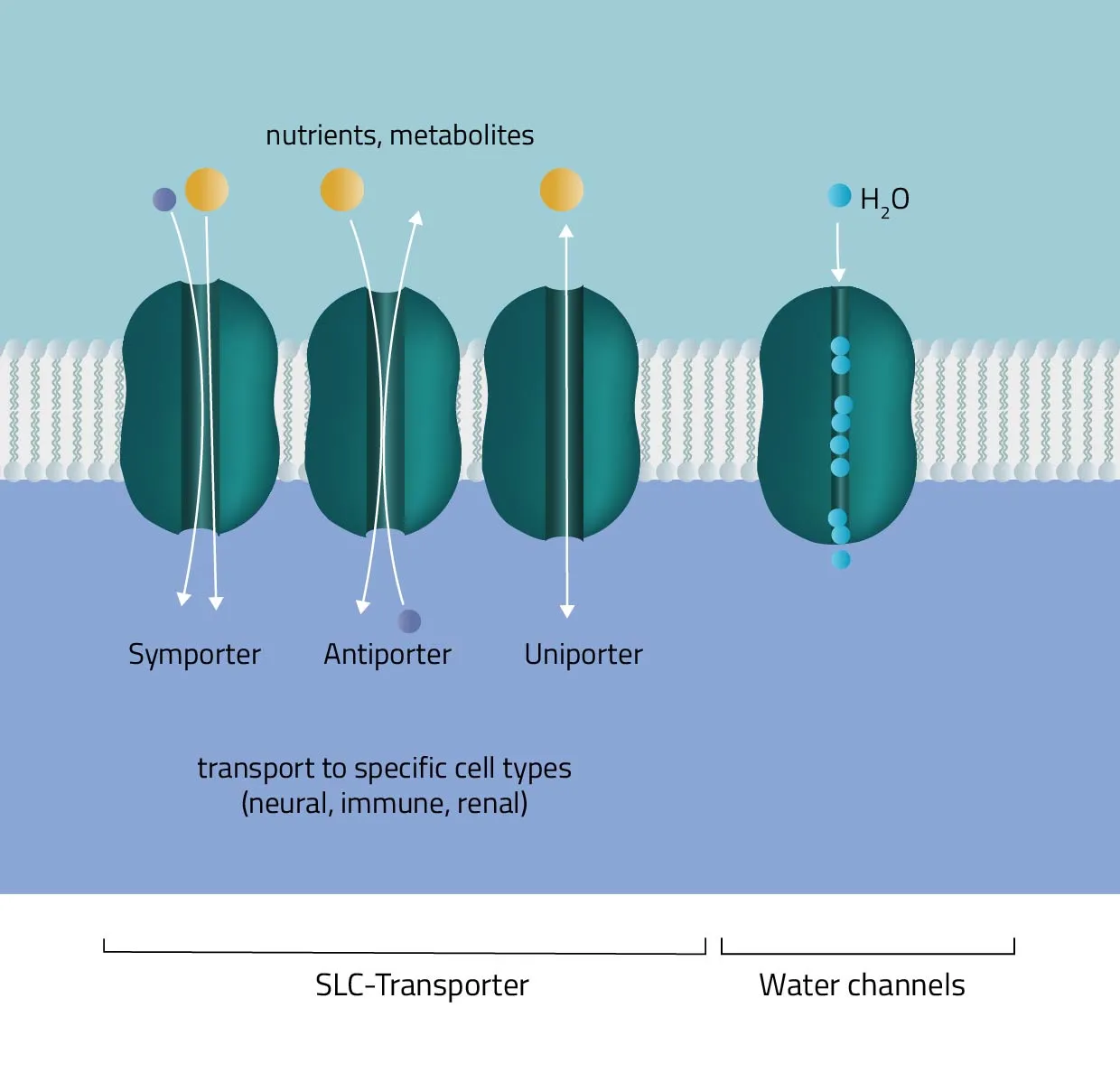
- ABC-Transporter
The ATP-binding cassette transporter superfamily is one of the largest gene families. In the majority of cases, they consist of multiple subunits, divided into hydrophobic transmembrane domains and hydrophilic membrane-associated ATPases. Functioning as molecular pumps they use the energy of ATP hydrolysis to move various solutes across cell membranes (Jones and George, 2002). - Ion channels
The pore-forming proteins facilitate the ion flow across cellular membranes. A subdivision according to types is made either by classification into gating mechanism, by ion type, or by cellular localization. A well-known type from the first group are voltage-gated ion channels, such as many Na+, K+, or Ca2+ channels. Biologically, they are a key component of the nervous system. Likewise, their involvement in muscle contraction or T-cell activation is equally important. - Membrane-bound ATPases
As indicated by its name, the ATPase superfamily utilizes ATP in order to perform its functions. They are categorized into F-, V-, and P-type ATPases. F- and V-type ATPases are classified as rotary ATPases, while ATPases of the P-type drive their conformational changes using free energy released by ATP hydrolysis (Palmgren and Nissen, 2011; Pizzagalli, Bensimon, and Superti-Furga, 2020). A well-known ATPase is the Na+/K+-exchanger, pumping sodium and potassium against their concentration gradients in order to maintain the cell membrane potential. - SLC-Transporter
Solute carrier proteins are a superfamily of transporter proteins, transferring a broad variety of solutes through membranes. These include molecules like sugar, amino acids, vitamins, or metals (Hediger et al., 2013). Therefore, they act as one of the main regulators of what enters or leaves a cell, and many physiological, as well as cellular processes, are depending on them (Pizzagalli, Bensimon, and Superti-Furga, 2020). - Water channels
Aquaporins or water channels facilitate the water flow across membranes. Thus, they play an elementary role in the maintenance of the water balance.
Functions
But not only do the transporter classes show a high diversity but the functions of the different classes are also undeniably complex. However, six overriding main functions are especially noteworthy:
- Enzymatic activity - procession of metabolites & substrates for various metabolic pathways
- Signal transduction - chemical messengers interact with membrane protein binding sites to send forth signals
- Transport (active/passive) - move molecules & other substances across the different cell membranes
- Cell-cell recognition - identification between cells, i.e. relevant for the immune system
- Intercellular joinings - different junctions, such as gap or tight junctions connect neighboring cells
- Anchorage / Attachment - important for cytoskeletal network, protein locations, and uphold of certain shapes
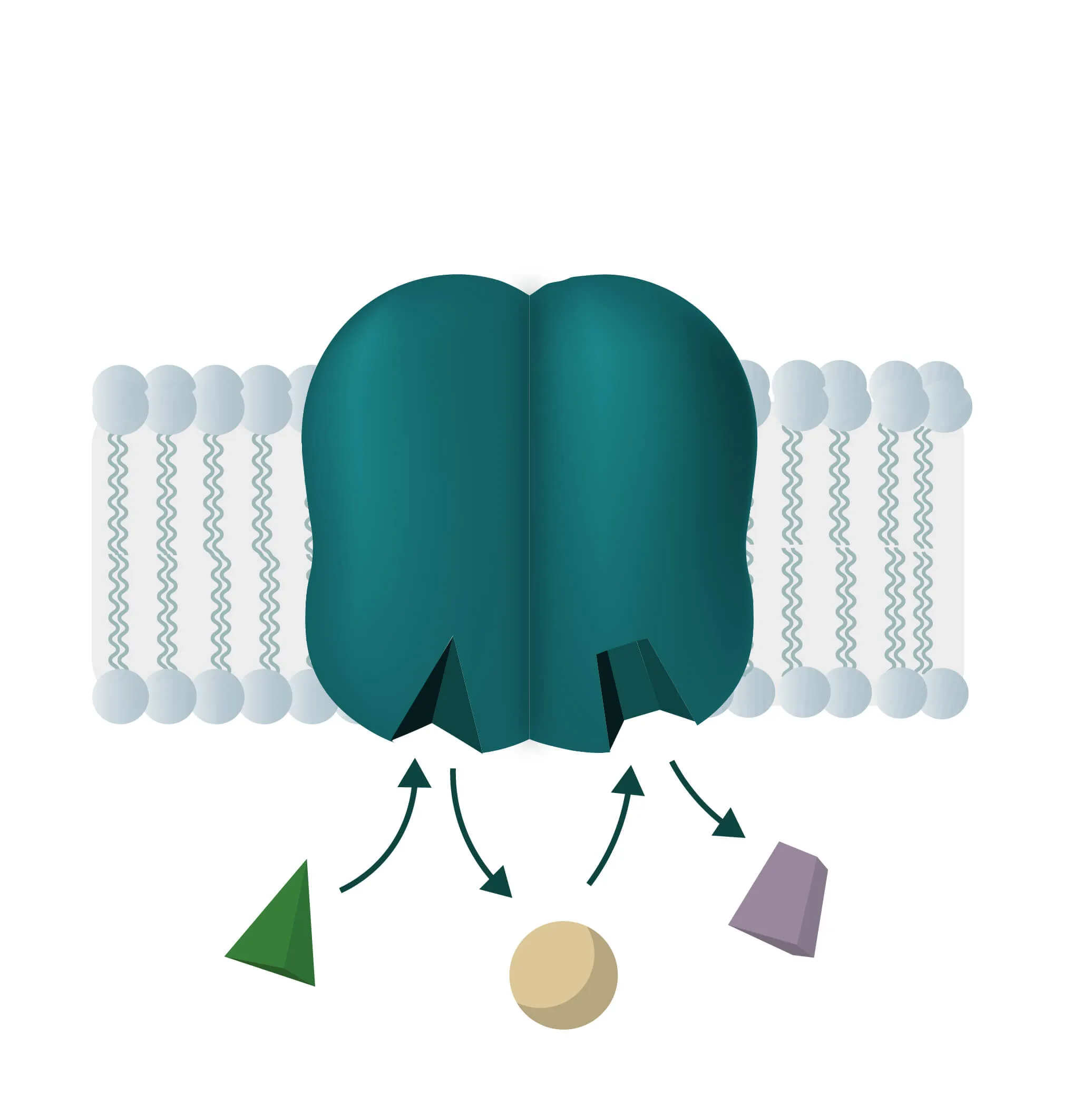
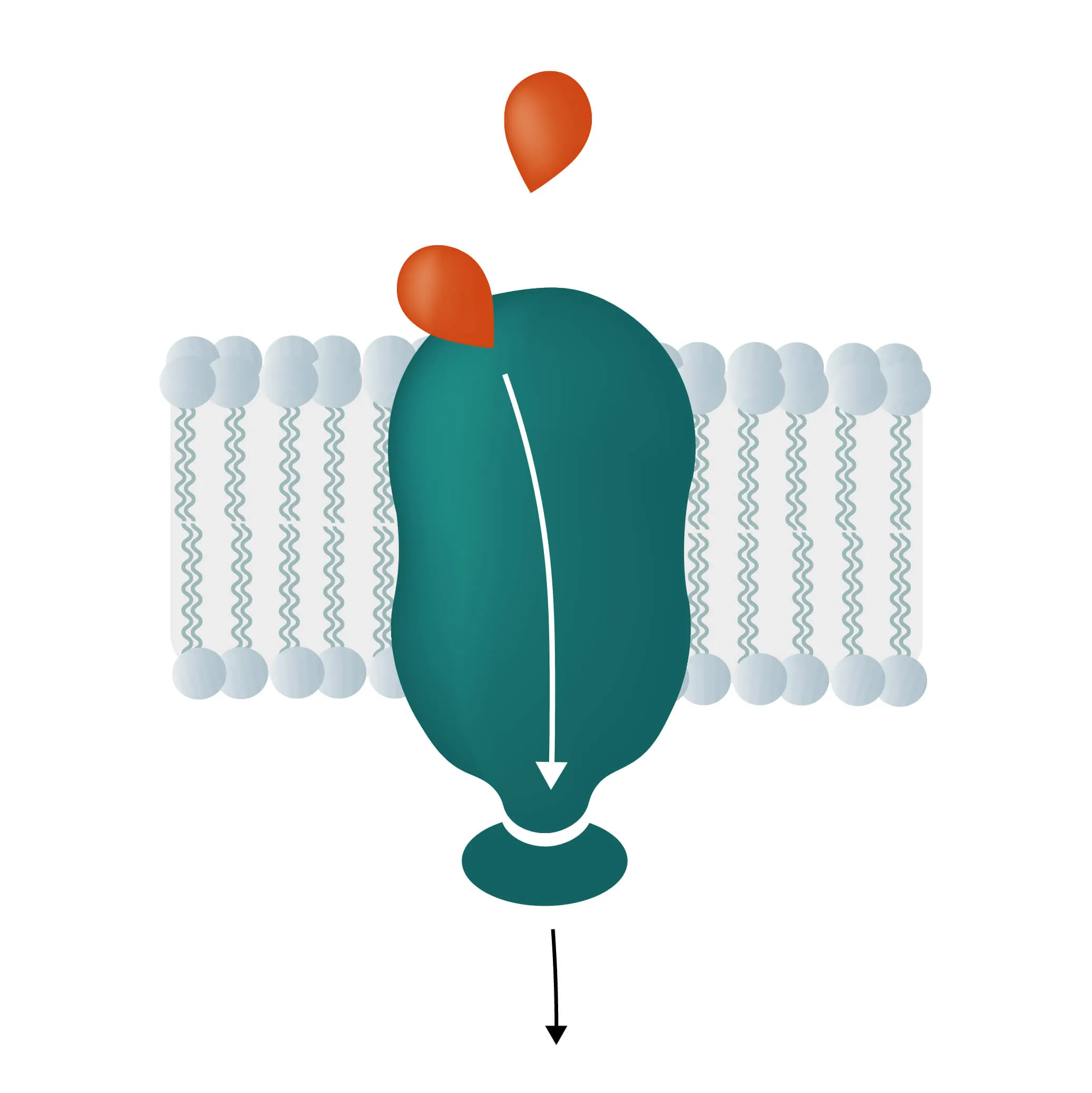
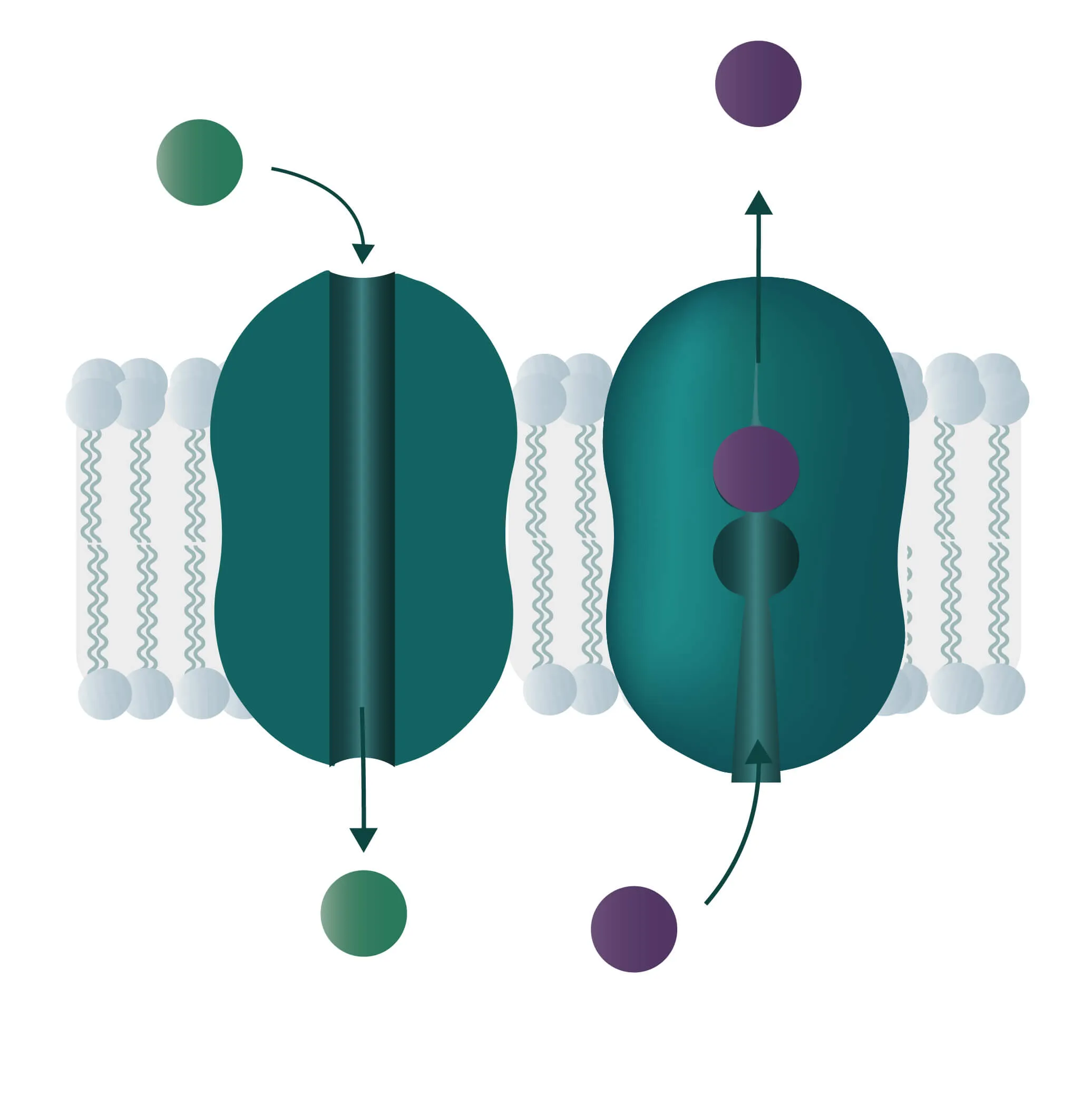

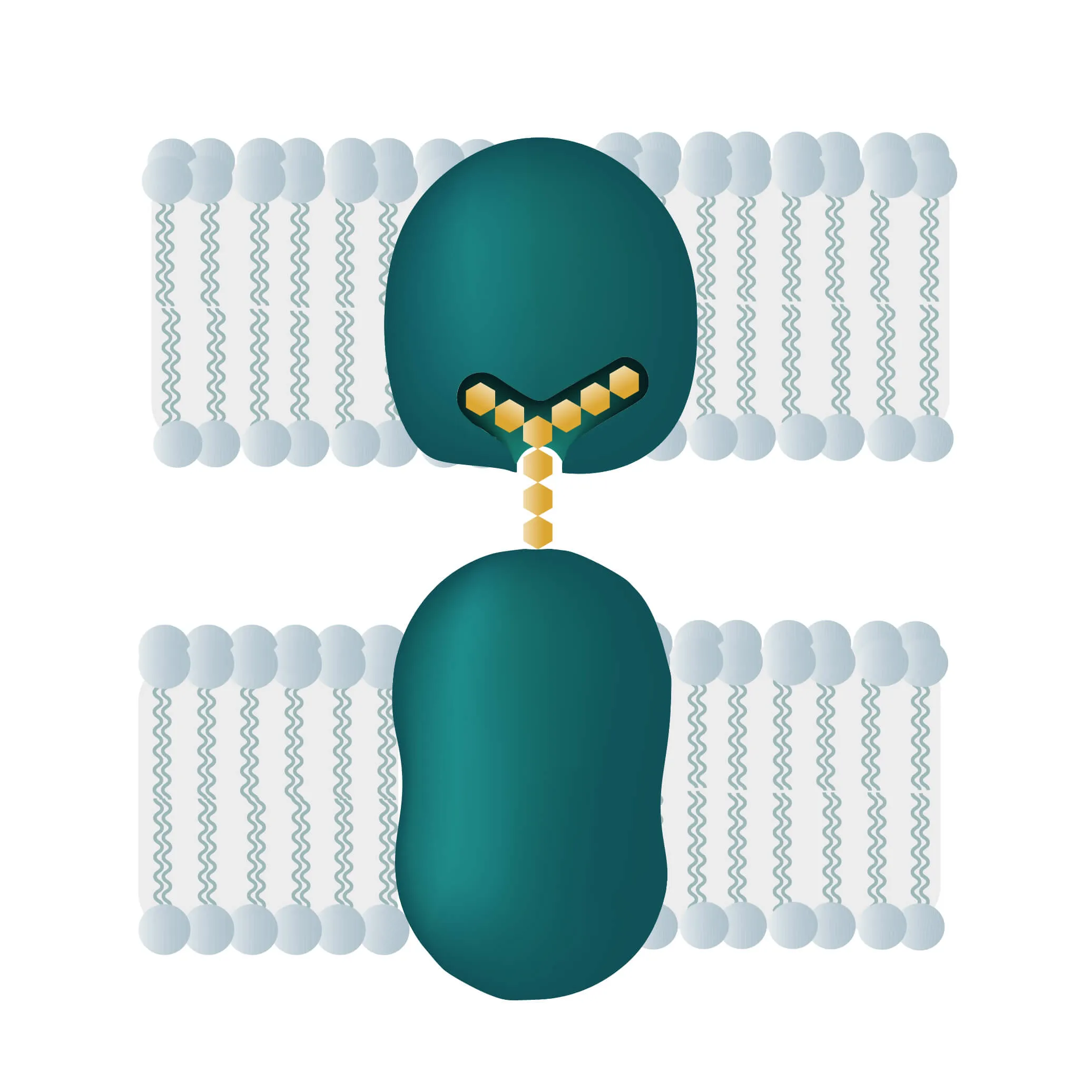
Composition, Structure, and Disposition
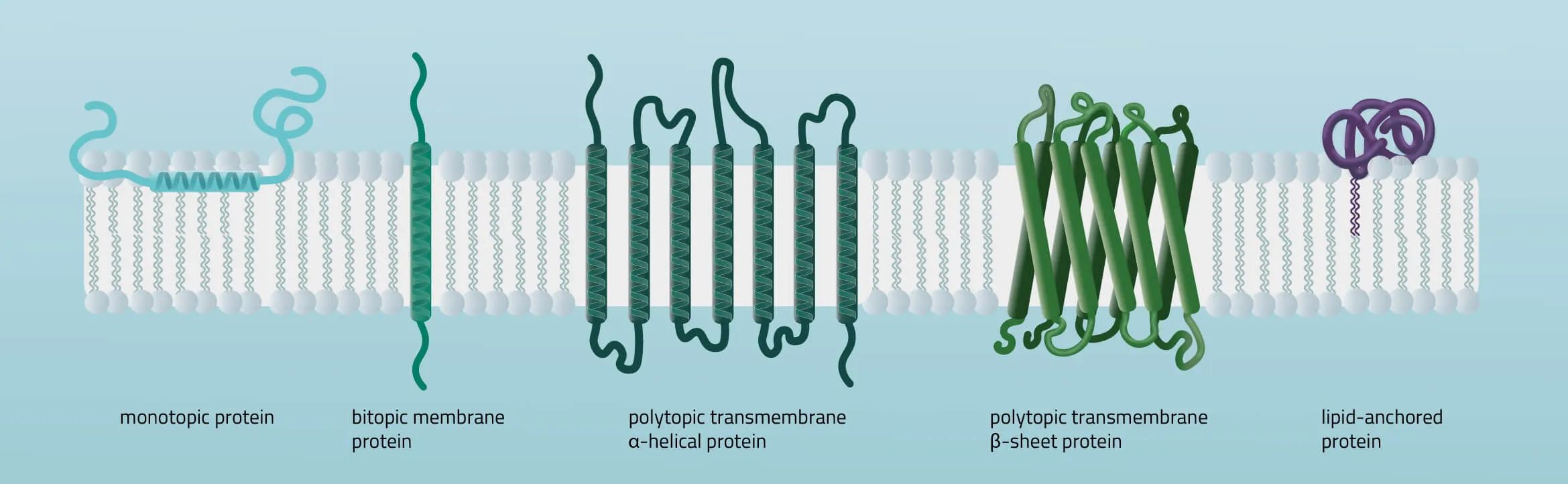
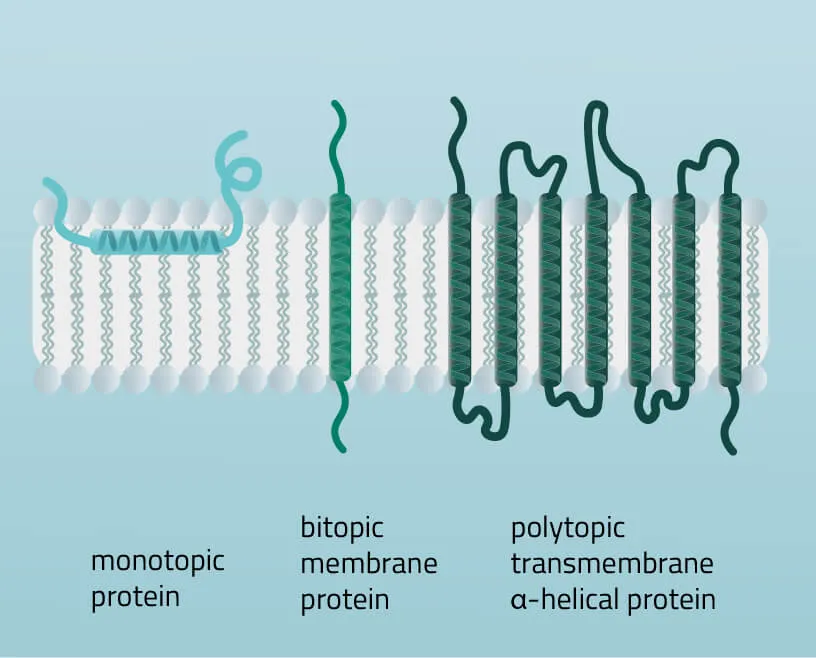
Membrane proteins vary in their composition and disposition. Different ratios of alpha-helical or beta-barrel structures will therefore entail different dispositions of those proteins. Structurally, membrane proteins are often amphipathic, having both hydrophilic and hydrophobic parts.
- Monotopic integral proteins are permanently attached to one side of the membrane bilayer. Interaction types involve for example an amphipathic α-helix parallel to the membrane plane (see Figure 11) or several hydrophobic loops anchoring the protein integrally.
- Bitopic integral proteins span the lipid bilayers only once. A typical bitopic structure consists of the transmembrane domain, plus the two cellular domains (extra and intra).
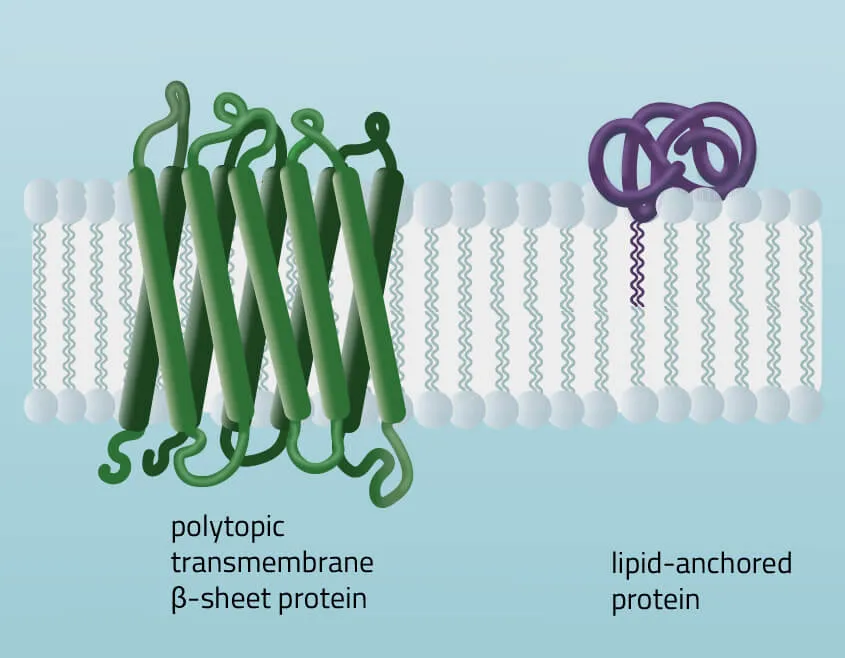
- Polytopic transmembrane proteins span the lipid bilayers more than once. Different compositions of alpha-helical and beta-barrel elements in the transmembrane domain are possible (see Figure 11 for two examples).
- Lipid-anchored proteins attach covalently to lipids embedded in the phospholipid bilayer. An interaction can take place by a covalent bond with a membrane lipid (lipidation) or by electrostatic/ionic interactions with membrane lipids.
As first proposed in 1972 by Singer and Nicholson the cellular membrane consists of a mosaic of components, labeled the fluid mosaic model (Singer and Nicolson, 1972). Phospholipids, cholesterol, membrane proteins, and carbohydrates as the main components give the membranes their fluid character. Integral proteins are only loosely attached to their surroundings, allowing slight movement inside the membrane.
Types of Membrane Transport Processes
In cell membranes, different kinds of channels, carriers, and pumps enable the transport of substances through the lipid bilayer. Many of them are highly specialized for specific interaction partners and only allow certain transports. In general, you can divide the transport processes into three broader categories diffusion, passive transport, and active transport.
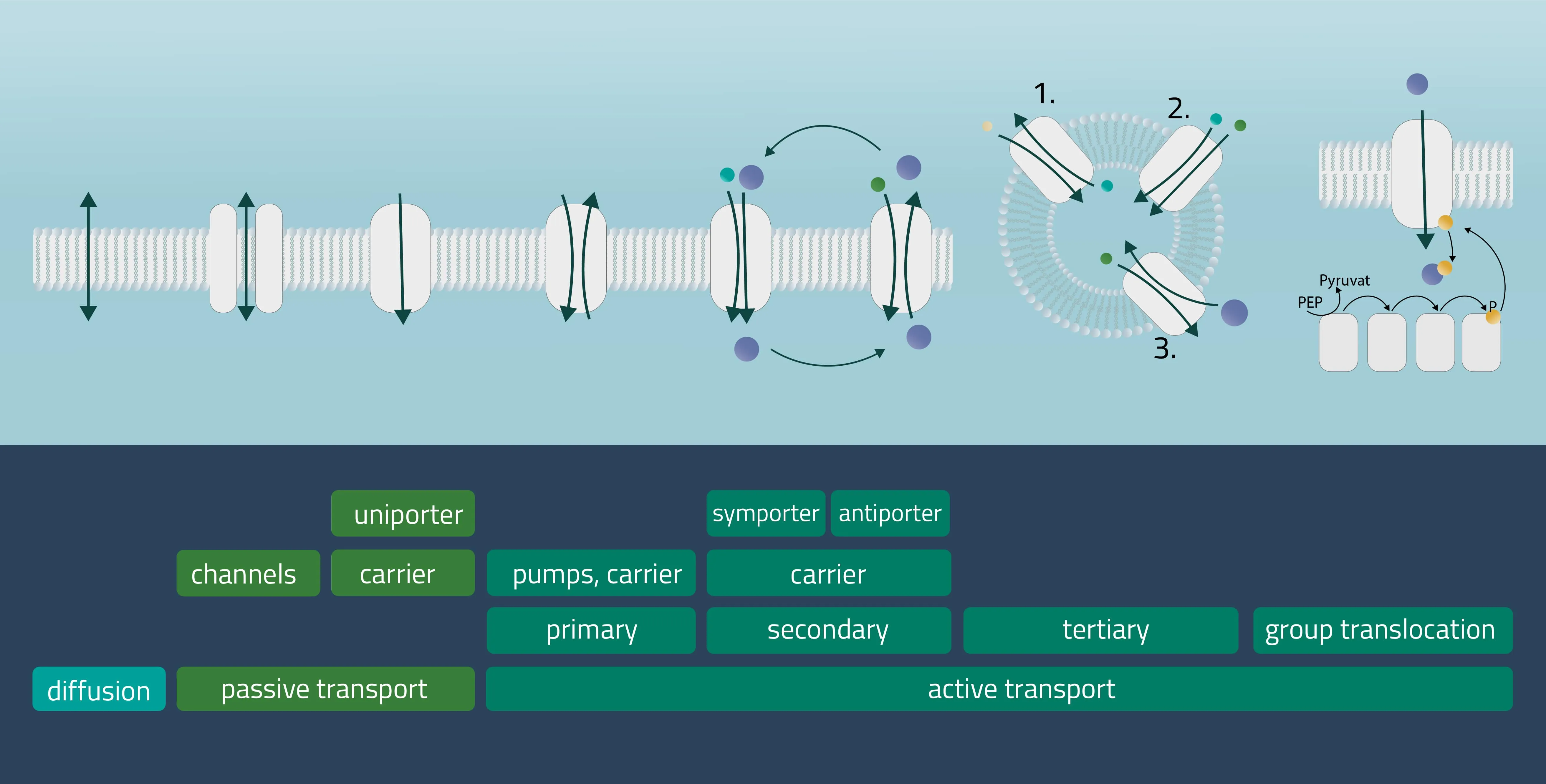
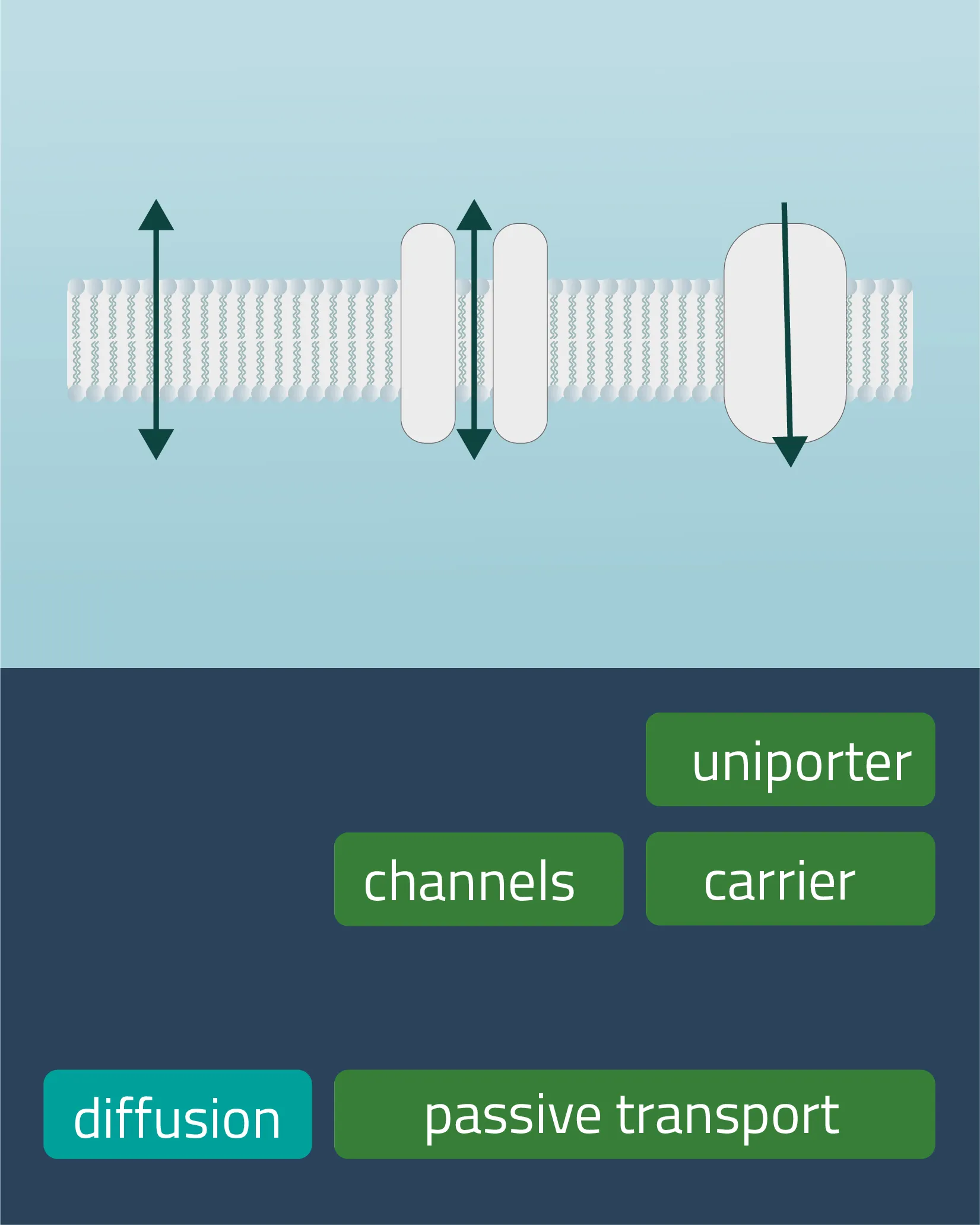
Movement of e.g. a molecule, ion, or particle from a region of a higher concentration, down its gradient, to a region of lower concentration. The movement continues until an equilibrium is reached. Charged particles can shift this membrane potential in one direction or the other.
Passive transport
Analogon of the simple diffusion for larger molecules, such as sugars or amino acids. Membrane transport proteins enable movement down a concentration gradient for substrates that under normal conditions are not able to pass the membrane.
- Channel proteins
Specialized transmembrane proteins spanning the cellular membrane. The gating mechanism can be triggered by ligands, changes in membrane potential, or by mechanical processes (e.g cytoskeletal changes). - Carrier proteins
For this type of passive transport, the molecule is transferred through conformational changes of a specialized carrier. The change is triggered by a substrate docking to the carrier. A means of transport can take place with a single molecule (Uniport), with two molecules moving in the same direction (Symport) or in opposite directions (Antiport).
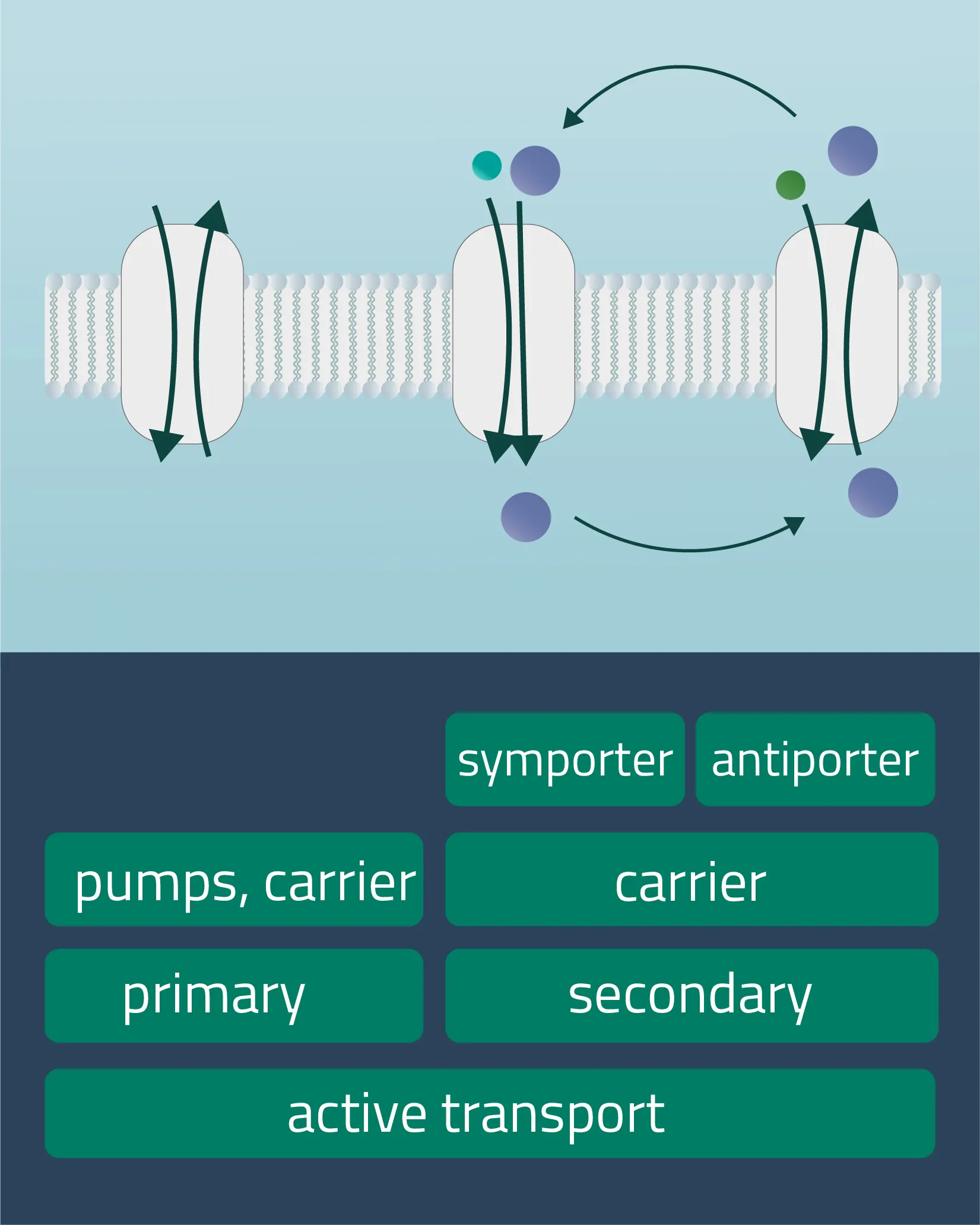
Active transport is a form of transport requiring energy from the outside in order to perform its mechanism. This enables the transport of molecules or ions against their concentration gradients or against electrical potential gradients. Forms of energy can be of chemical nature (ATP) or electric charge. It is also possible to utilize a concentration gradient as an energy source.
- Primary active transport
Protons and inorganic ions are moved through the cellular membrane by harnessing the energy of ATP. The Na-K-Pump is an example of this transport form, transferring three positively charged sodium and two likewise positively charged potassium ions. - Secondary active transport
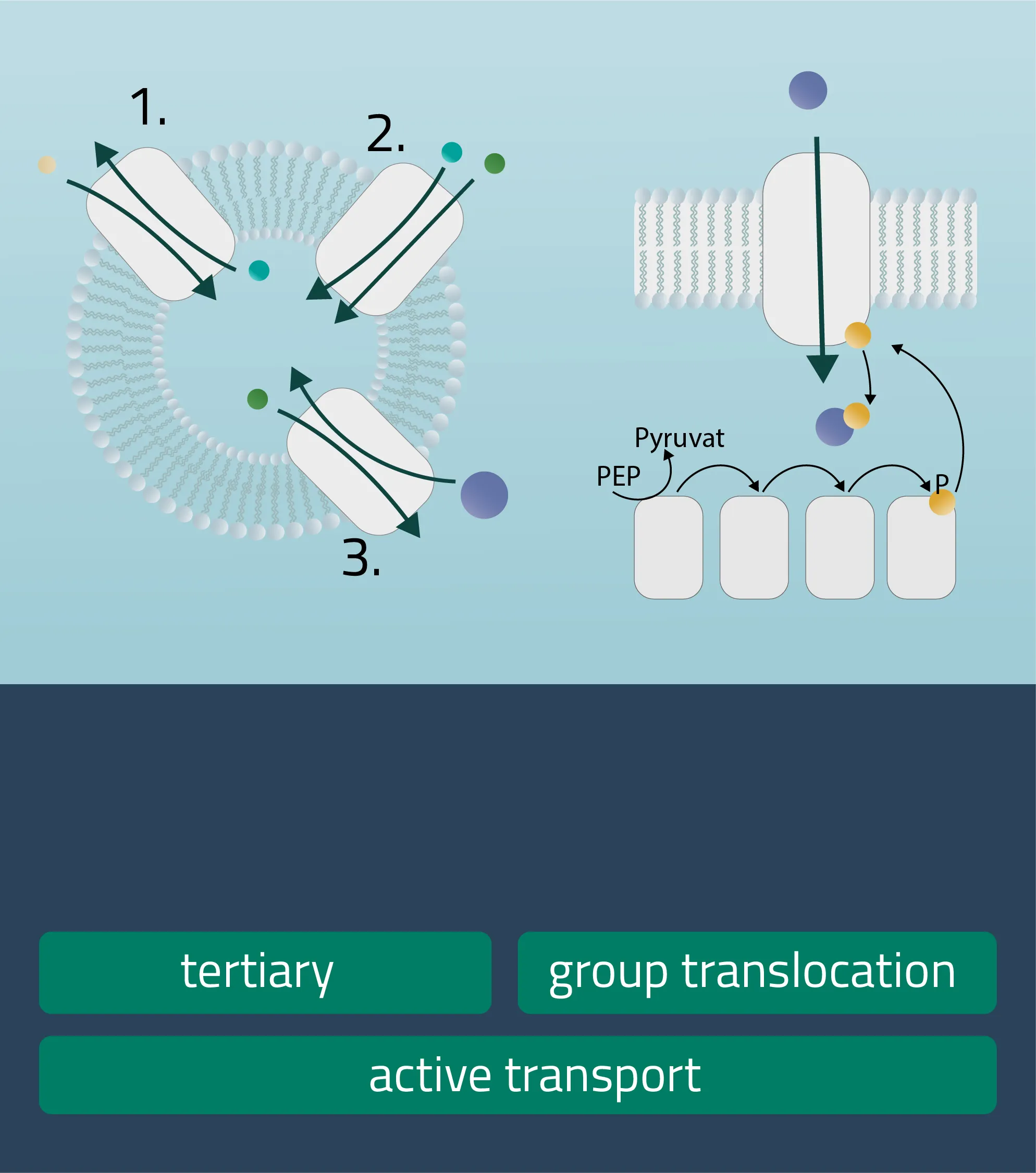
Analog to the passive carrier protein sym- and antiporters, this time one of the ions moves with its concentration gradient while the second one moves against it. This can be achieved in the same direction (Symport) or in opposite directions (Antiport). Therefore, an electrochemical gradient is a driving force here. - Tertiary active transport
For this type, three transporters need to function in a series. A first transporter establishes an electrochemical gradient for molecule A (Primary active). The second transporter utilizes molecule A to establish a favorable electrochemical gradient for molecule B (Secondary active). Finally, transporter three uses the gradient of molecule B to move molecule C against its concentration gradient (Hamm, Alpern, and Preisig, 2008). - Group translocation
This is a special transport form of bacteria. The substrate to be transported is chemically altered in the process. As a result, no concentration gradient is created. As an energy form ATP can be utilized, but there are other energy sources like PEP (Phosphotransferase system), utilized for glucose transport processes as well (see Figure 12, group translocation).
Historical Relevance
Just over 50 years have passed since the early beginnings of modern, molecular membrane biology in the 1960s to 1970s. Discoveries such as the deciphering of the molecular cell membrane structure or their underlying general mechanisms have paved the way for many important insights and fields of application in modern medicine. Meanwhile, the characterization of membrane-bound proteins faced even greater hurdles. Therefore, it took until 1978, five years after the postulation of the fluid mosaic model, to report the first successful characterization of an integral membrane protein. This was human glycophorin obtained from the cell membrane of erythrocytes (Singer, 2004).
Since those early days of membrane protein research, many milestones have been achieved. In medicine, in particular, the knowledge gained has been used in a variety of ways to improve and combat numerous clinical conditions.
Challenges and Future Prospects for Medicine / Industry
Cell membranes act as the final gatekeepers for many substrates and drugs. Highly specialized membrane proteins, therefore, interact in a fraction of a second with a plethora of substances to determine which of them are allowed through and which are not. About half of all currently approved drugs act on this protein type as a therapeutic target structure to cause signal transductions, trigger cascades, or catalyze reactions in order to combat various clinical conditions (Baker, 2010).
In spite of this, little is still known about many membrane protein structures and their underlying working principles, due to their challenging solubilization criteria. Modern medicine would greatly benefit from a deeper understanding of this protein type. Recently though, this old problem is picking up speed again, as various AI systems like DeepMind's AlphaFold2 join the fight. They are capable of predicting a 3D protein structure directly from its amino acid sequence with ever-increasing accuracies.
Meanwhile, experimental methods already being applied like the nanodisc systems, cycloalkane-modified amphipols, or AASTY copolymers will develop further as well, pushing our boundaries of knowledge forward.
In the future, the intertwinement of AI and experimental methods like these will introduce a new era of biological and medical research, shaping fields like digital biology or biomedical computing. Novel approaches will renew our drug discovery processes, accelerating them immensely. Whatever path we pursue, the membrane proteome will play a vital part in it.
In the future, the intertwinement of AI and experimental methods like these will introduce a new era of biological and medical research, shaping fields like digital biology or biomedical computing. Novel approaches will renew our drug discovery processes, accelerating them immensely. Whatever path we pursue, the membrane proteome will play a vital part in it.
Literature
- Piovesan, A., Antonaros, F., Vitale, L. et al. "Human protein-coding genes and gene feature statistics in 2019." BMC Res Notes 12, 315 (2019). https://doi.org/10.1186/s13104-019-4343-8
- Baker, M. "Making membrane proteins for structures: a trillion tiny tweaks." Nat Methods 7, 429–434 (2010). https://doi.org/10.1038/nmeth0610-429
- Salzberg S. L. "Open questions: How many genes do we have?." BMC biology, 16(1), 94.(2018). https://doi.org/10.1186/s12915-018-0564-x
- Singer, S. J. “Some early history of membrane molecular biology.” Annual review of physiology vol. 66 (2004): 1-27. doi:10.1146/annurev.physiol.66.032902.131835
- Jones, P.M., George, A.M. "Mechanism of ABC transporters: A molecular dynamics simulation of a well characterized nucleotide-binding subunit." PNAS 99 20 (2002) 12639-12644; https://doi.org/10.1073/pnas.152439599
- Hediger, M.A., Clémençon, B., Burrier, R.E. and Bruford, E.A. "The ABCs of membrane transporters in health and disease (SLC series): introduction." Mol Aspects Med 34, 95–107. (2013)
- Pizzagalli, M.D, Bensimon, A., Superti-Furga, G. "A guide to plasma membrane solute carrier proteins." The FEBS Journal, vol. 288 9, (2021) 2784-2835; https://doi.org/10.1111/febs.15531
- Palmgren, M.G. and Nissen, P. “P-type ATPases.” Annual review of biophysics vol. 40 (2011): 243-66. doi:10.1146/annurev.biophys.093008.131331
- Hamm, L.L., Alpern R.J., Preisig, P.A. "Chapter 54 - Cellular Mechanisms of Renal Tubular Acidification." Seldin and Giebisch's The Kidney (Fourth Edition), Academic Press, 2008, Pages 1539-1585, ISBN 9780120884889, https://doi.org/10.1016/B978-012088488-9.50057-7.
- Singer, S.J., Nicolson, G.L. “The fluid mosaic model of the structure of cell membranes.” Science (New York, N.Y.) vol. 175,4023 (1972): 720-31. doi:10.1126/science.175.4023.720
- Yeagle, P.L. "Chapter 10 - Membrane Proteins" The Membranes of Cells (Third Edition), Academic Press, 2016, Pages 219-268, ISBN 9780128000472, https://doi.org/10.1016/B978-0-12-800047-2.00010-3.
