Cell-Free Expression
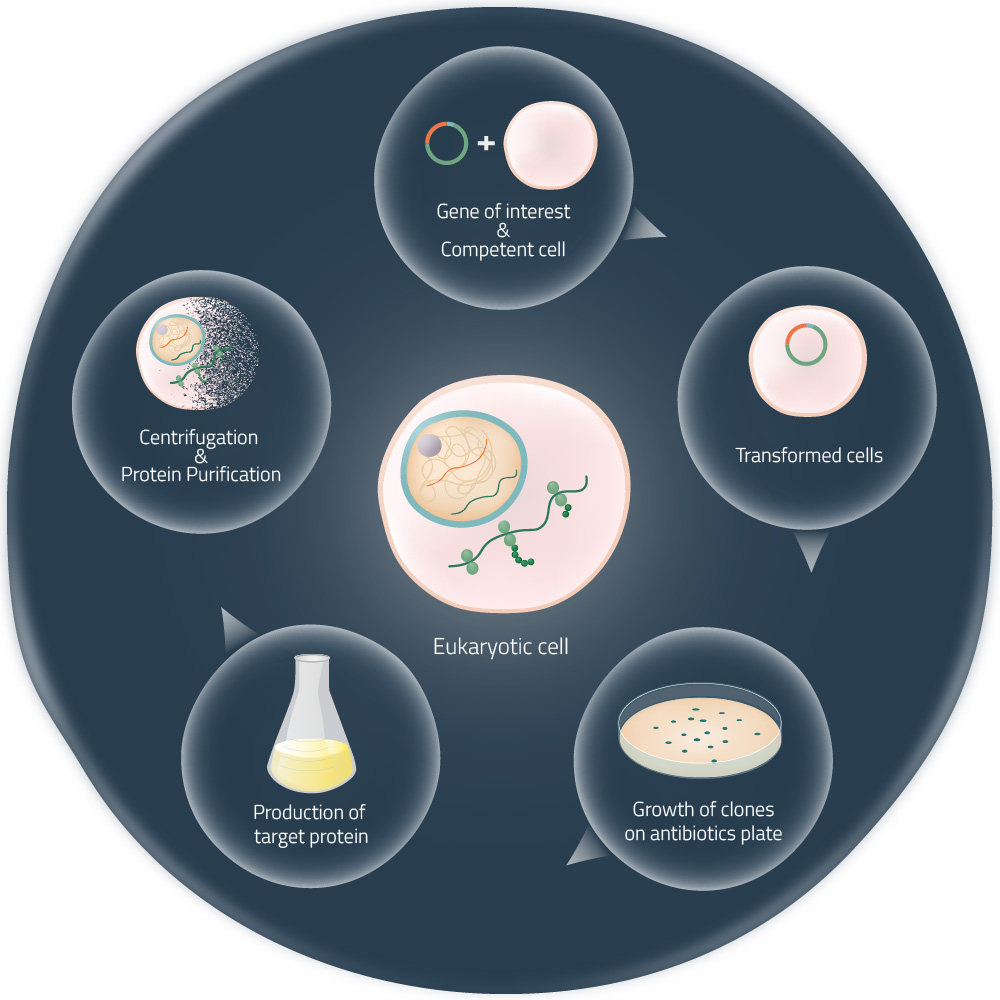
Cell-free expression systems (or cell-free protein synthesis) are alternative approaches to the production of target proteins, without the need for living cells (see Fig. 2). They are generated by the lysis of eukaryotic or bacterial cells and the consequent removal of all components not required for protein expression. Thus, the received cell lysates can be managed very precisely. The addition of suitable DNA templates in combination with components like amino acids, NTPs, or certain cofactors (see Fig. 1) will, later on, start the in vitro protein expression.1
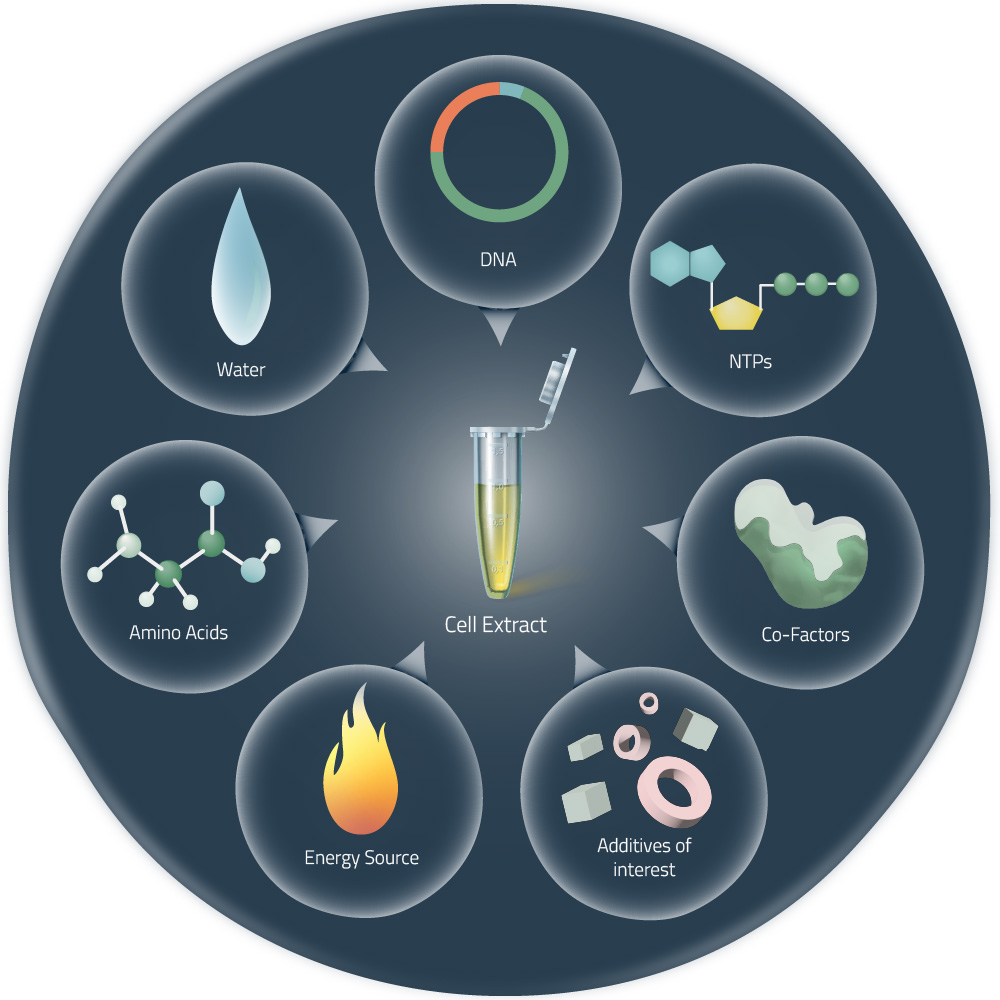
The Story of Cell-free Protein Expression
Historically, the technique has been around for roughly sixty years. Therefore, quite a few generations of scientists were able to make use of this method. Very notable is the discovery of the first codon, performed by Matthaei and Nirenberg in 1961. With the help of a cell-free system they were able to observe, that their synthesized polypeptide, solely consisting of a poly-uracil RNA sequence, only contained the amino acid phenylalanine.2
Recently, the attention is growing again as big players in the industry are taking interest in it. Well-established methods to produce a target protein are often time-consuming and require living cells (see Fig. 2). A broader and easier industrial production of biomolecules, on the basis of cell-free protein expression, will enable many companies to produce more cost- and resource-efficient in the future (e.g. Fraunhofer - Lighthouse Project »Cell-Free Bioproduction«).
Recently, the attention is growing again as big players in the industry are taking interest in it. Well-established methods to produce a target protein are often time-consuming and require living cells (see Fig. 2). A broader and easier industrial production of biomolecules, on the basis of cell-free protein expression, will enable many companies to produce more cost- and resource-efficient in the future (e.g. Fraunhofer - Lighthouse Project »Cell-Free Bioproduction«).
Benefits and Drawbacks
Cell-free protein synthesis offers quite a few advantages in comparison to conventional in vivo methods. On the other hand, it also has its pitfalls that need to be considered when deciding on cell-free systems. To give you a general overview of what to expect we listed a couple of benefits and drawbacks here:

- Cell-free reactions are easy to set up and take only several hours to express your protein. Optimization of reaction conditions can be done quickly and parallel by the use of small-scale reactions (50-100µl).
- Small handling volumes: mg amounts of soluble proteins obtainable from only a few ml of reaction volume.
- Open system allows addition of many components (natural or unnatural) to the reaction, e. g. reducing/oxidizing elements, chaperones, labeled amino acids, or detergents. Direct manipulation of the chemical environment is possible.
- Proteins can be utilized in applications like nuclear magnetic resonance (NMR)
- Reconstitution of membrane proteins into nanodisc systems in their nascent state3-5
- Eukaryotic cell-free lysates provide post-translational modifications such as core glycosylation or phosphorylation and natural membrane components (e.g. microsomes) for membrane protein insertion.6
- Aspects like toxicity7, protease digestion, or DNA template instability are of no concern since the cells are not alive anymore and the responsible cellular components have been removed.
- Proteins and complexes with a multimeric conformation can be expressed and it is possible to affect the ratios of their respective subunits by DNA template titration.

A cell-free expression can turn into a costly process when larger quantities of protein are needed or if your proteins only show poor expression. Therefore, it is advised to assess achievable yields for each protein beforehand in small-scale experiments to evaluate the increased cost for upscaled experiments. This is especially true for more complex cell-free lysates from eukaryotic sources.
Another challenge for many researchers are endogenous nucleases. Endo- and exonucleases can quickly degrade a DNA of interest, leading to significantly reduced yields. Due to their circular shape, plasmids are not affected by exonucleases. Linear expression templates however are very vulnerable to endogenous nucleases. A possible countermeasure for this can be the bacteriophage λ -gam protein. Gam is able to inhibit the exonuclease RecBCD, usually found in E. coli.8
Another challenge for many researchers are endogenous nucleases. Endo- and exonucleases can quickly degrade a DNA of interest, leading to significantly reduced yields. Due to their circular shape, plasmids are not affected by exonucleases. Linear expression templates however are very vulnerable to endogenous nucleases. A possible countermeasure for this can be the bacteriophage λ -gam protein. Gam is able to inhibit the exonuclease RecBCD, usually found in E. coli.8
Table 1: Comparison of four different cell-free expression systems
| E.coli | Wheat germ | Insect | Mammalian | |
|---|---|---|---|---|
| Yield (relative) | high (several mg/ml) | medium | medium-low | low |
| Cost (relative) | low | medium | medium | high |
| Phosphorylation | no | yes | yes | yes |
| Glycosylation | no | core | core | yes (mammalian) |
| Membrane protein options | insoluble detergent nanodisc liposomes | detergent liposomes |
|
|
| Commercially available |
|
|
batch Mmix | batch Mmix |
The four cell-free expression systems all have their respective application areas in which they perform best. E. coli for example yields high amounts of protein and is relatively tolerant to additives, but performs rather badly with eukaryotic proteins. Wheat germ extracts or insect cell systems, like Sf219, allow the translation of larger proteins in combination with moderate yields. Lastly, mammalian expression systems from rabbit reticulocyte lysates enable the user to perform a cap-independent translation while utilizing a eukaryotic system. Drawbacks however are lower relative yields or increased sensitivity to additives.
What is Cell-free Protein Synthesis used for?
Because proteins are expressed in an open system, there are many applications that are facilitated by cell-free protein synthesis in comparison to expression in living cells. These include:
Applications
- Expression screening: With cell-free expression, you can go from DNA template - or even multiple reactions in parallel - to Western Blot result in two days, making the system attractive for fast screening of multiple constructs and reaction conditions.
- Protein labeling: The open system can be combined with labeled amino acids to introduce e.g. biotin, fluorescent labels, or modified/unnatural amino acids.
- NMR: Isotope-labeled amino acids can be easily added to the mix to obtain proteins ready for NMR.
- Crystallization / Biophysical: In combination with selenomethionine, protein-heavy metal derivatives are obtained in a straightforward procedure; expression of proteins for structure-function analysis
- Further applications: mutagenesis studies, production of toxic proteins or enzyme screening
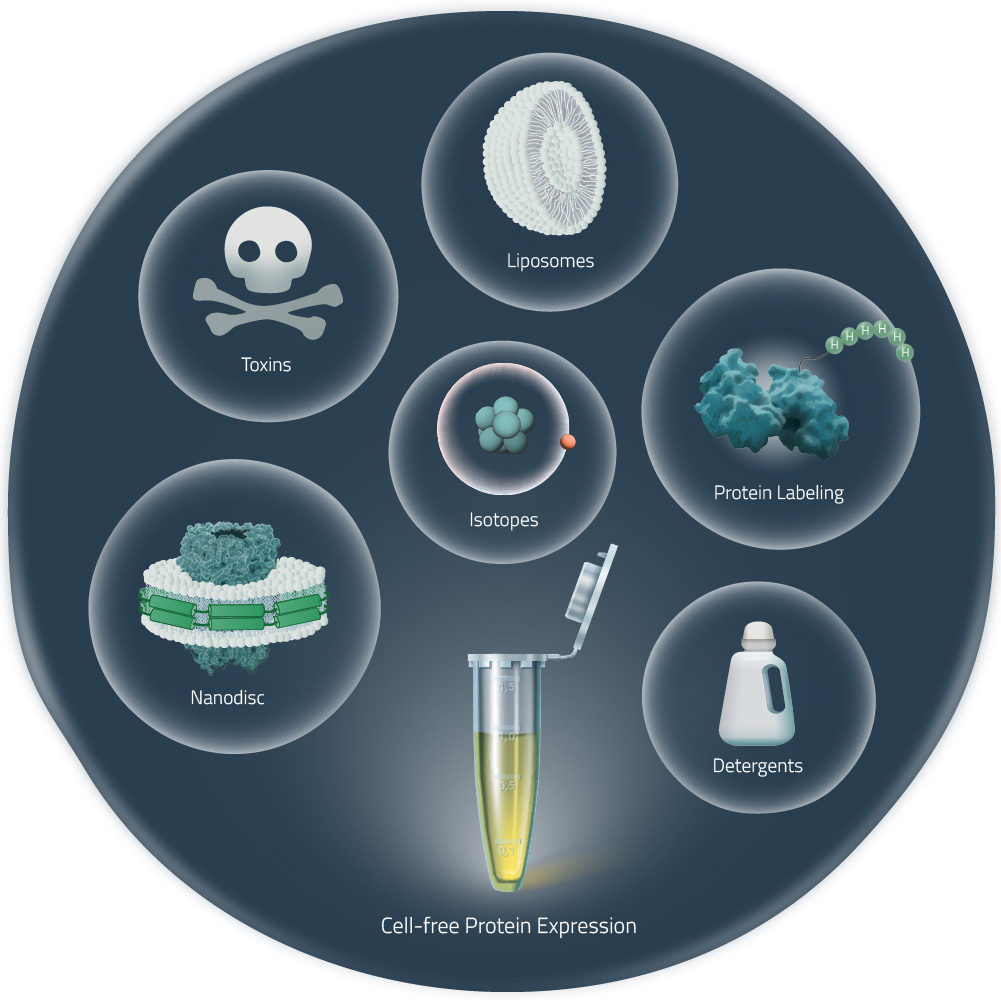
Cell-free expression of membrane proteins
- Insoluble expression: without the addition of any kind of lipid or detergent, membrane proteins form aggregates that in many cases can be refolded into functional proteins.
- Detergent: Detergents, especially of the Brij series, have been added in many cases to solubilize membrane proteins. If needed, a detergent exchange can be performed in subsequent steps, preferably during purification.10-15
- MSP-Nanodiscs: Adding MSP-nanodiscs to cell-free reactions is the simplest and most elegant way to obtain stabilized proteins that can be used for a variety of assays without the need for detergent.3-5, 16-18 Applications range from biophysical assays like SPR (surface plasmon resonance spectroscopy), NMR or mass spectrometry to cryo-EM. Recently, there is evidence that membrane proteins expressed in cell-free lysates in the presence of nanodiscs can be crystallized by in meso methods.17 Learn more about nanodiscs.
- Liposomes and lipid particles: Liposomes, microsomes, or lipid-containing structures such as bicelles have been added to cell-free reactions, particularly in combination with insect cell lysates, for membrane protein expression.6 Using appropriate detergents, membrane proteins can be shifted from nanodiscs into bicelles.16
Batch vs. CECF methods
Cell-free reactions can be separated into two possible procedures:
- Batch: Batch cell-free reactions are very easy to set up in simple vials like microcentrifuge tubes. Reaction times are short (1 - 3 hours) because energy components are used up quickly, and the accumulation of end products slows down the enzymatic reactions. Therefore, protein yields are typically low.
- CECF or dialysis: Continuous exchange cell-free (CECF) reactions are set up in dialysis chambers so that energy components are fed into the reaction, and end products such as ADP are removed. This way, reaction times up to 24 hours are possible, and the protein yields obtained can reach several mg per ml of reaction for well-expressing soluble proteins.19
References:
- Endo, Y. et al. (eds) Cell-Free Protein Production, Methods and Protocols. Methods in Molecular Biology vol. 607 (2010)
- Nirenberg, M. W., Matthaei, J. H. "The Dependence of Cell- Free Protein Synthesis in E. Coli Upon Naturally Occurring or Synthetic Polyribonucleotides". Proceedings of the National Academy of Sciences of the United States of America. 47 (10): 1588–1602 (1961)
- Roos, C. et al. Characterization of co-translationally formed nanodisc complexes with small multidrug transporters, proteorhodopsin and with the E.coli MraY translocase. Biochim. Biophys. Acta 1818; 3098-3106 (2012)
- Proverbio, D. et al. Functional properties of cell-free expressed human endothelin A and endothelin B receptors in artificial membrane environments. Biochim. Biophys. Acta 1828, 2182-92 (2013)
- Roos, C. et al. High-level cell-free production of membrane proteins with nanodiscs. In: Cell-free Protein Synthesis: Methods and Protocols. Alexandrov, K., and Johnston, W.A. (eds), Methods in Molecular Biology vol. 1118, Chapter 7 (2014)
- Sachse, R. et al. Membrane protein synthesis in cell-free systems: From bio-mimetic systems to bio-membranes. FEBS Lett. 588,2774-2781 (2014)
- Jackson, A.M. "Cell-free protein synthesis for proteomics". Briefings in Functional Genomics and Proteomics. 2 (4): 308–319 (2004)
- Court, R., Cook, N., Saikrishnan, K., Wigley, D. The Crystal Structure of λ-Gam Protein Suggests a Model for RecBCD Inhibition, Journal of Molecular Biology, Volume 371, Issue 1 (2007)
- Suzuki, T. et al. An insect cell-free system for recombinant protein expression using cDNA resources. Methods in molecular biology vol. 577 (2009): 97-108.
- Boland, C. et al. Cell-free expression an in meso crystallization of an integral membrane kinase for structure determination. Cell. Mol. Life Sci (2014)
- Hein, C. et al. Hydrophobic environments in cell-free systems: Designing artificial environments for membrane proteins. J. Eng. Life Sci. 14, 365-79 (2014)
- Proverbio, D. et al. Membrane protein quality control in cell-free expression sysems: Tools, strategies and case studies. Membrane proteins production for structural analysis (Isabelle Mus-Veteau, ed.) Springer Heidelberg. ISBN:978-1-4939-0662-8.
- Haberstock, S. et al. A systematic approach to increase the efficiency of membrane protein production in cell-free expression systems. Prot. Exp. Purific. 82, 308-16 (2012)
- Matthies, D. et al. Cell-free expression and assembly of a macromolecular membrane protein complex. J. Mol. Biol. 413, 593-603 (2011)
- Schneider, B. et al. Membrane protein expression in cell-free systems. Metho. Mol. Biol. 601, 165-86 (2010)
- Laguerre, A. et al. From nanodiscs to isotropic bicelles: A procedure for solution nuclear magnetic resonance studies of detergent-sensitive integral membrane proteins. Structure 24, 1-12 (2016)
- Nikolaev, M. et al. Integral membrane proteins can be crystallized directly from nanodiscs. Cryst. Growth Des. 17(3), 945–948 (2017)
- Henrich, E. et al. Analyzing native membrane protein assembly in nanodiscs by combined non-covalent mass spectrometry and synthetic biology. eLife 6:e20954 (2017)
- Schwarz, D. et al. Preparative scale expression of membrane proteins in E.coli based continuous exchange cell-free systems. Nat. Protocols 2, 2945-57 (2007)


