What Are Nanodiscs - Copolymers
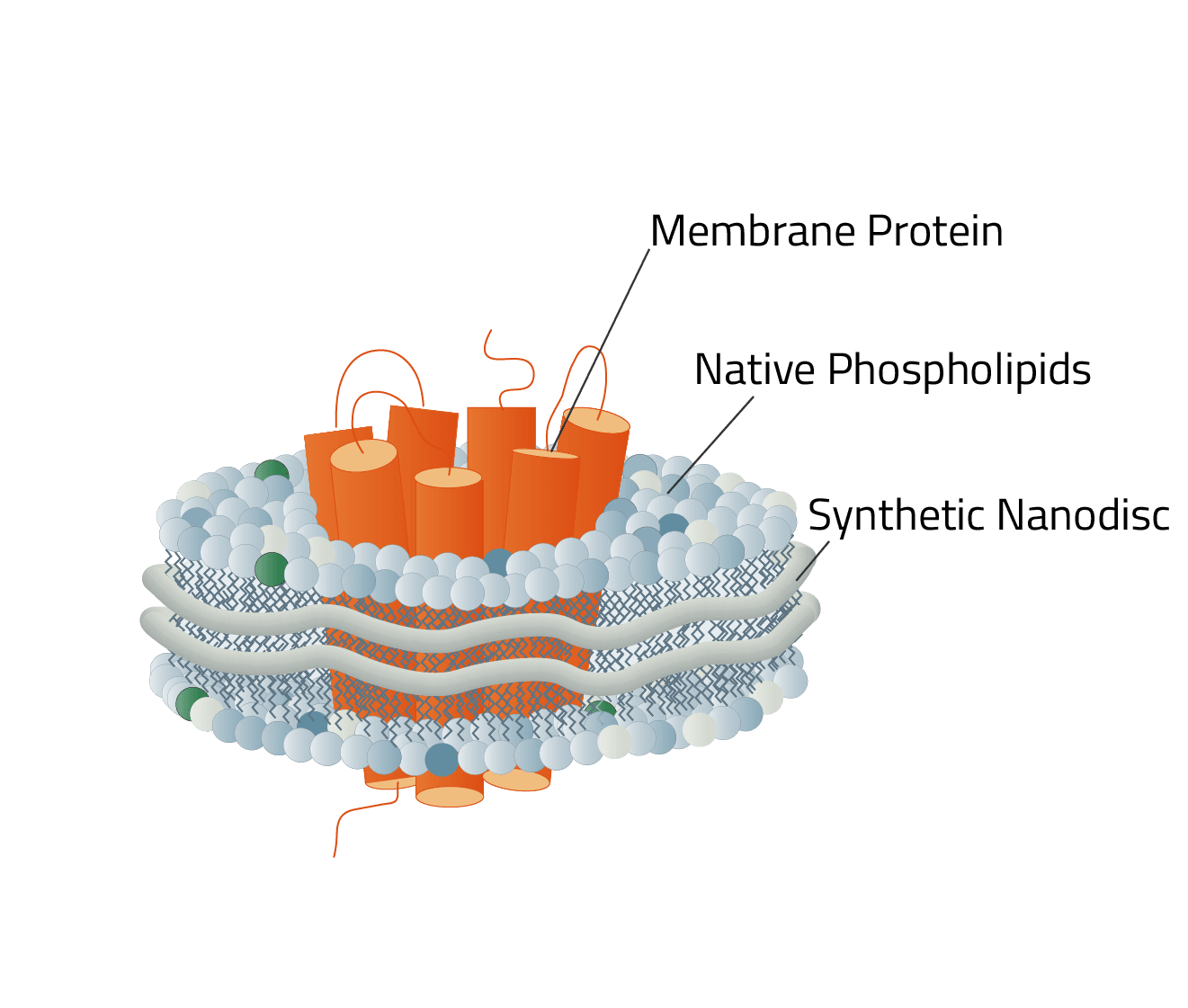
Copolymer Nanodisc are small disc-shaped structures, composed of cell membrane phospholipids that are held together by a Copolymer ring.
They provide a mobile, near identical copy, of the native membrane protein environment in the cell membrane. Thus, they circumvent the problems of solubilization detergents and allow us to stabilize and isolate membrane proteins in their active state for further scientific research.
There are several different Copolymers that can be used to create Copolymer Nanodisc. Each with its own advantages and disadvantages
How does a Copolymers form a Nanodisc for Membrane Proteins?
Cell membranes provide the environment for one of our most important protein groups: the membrane proteome. Being divided into peripheral and integral proteins, they both share a certain hydrophobicity of some sort, impeding solubilization under normal conditions.
The problem is that when these hydrophobic regions come into contact with water or another hydrophilic medium, the 3D structure of the membrane protein collapses and consequently loses its functionality.
To avoid this from happening, protein scientists have traditionally used detergents to cover up those vulnerable parts of the membrane protein. But detergent based approaches come with their own sets of problems, like time-consuming screening processes or 3D structure interferences.
Copolymers however are able to form a Copolymer chain out of their monomers to insert into a cell membrane surrounding your desired membrane protein (see Fig. 2, red thread at the top). Like a cookie cutter, the membrane protein is dissolved from the membrane and the Copolymer keeps the membrane protein stable in the newly formed nanodisc.
Consequential affinity chromatography steps allow for precise isolation of your stabilized target protein and further scientific analysis.

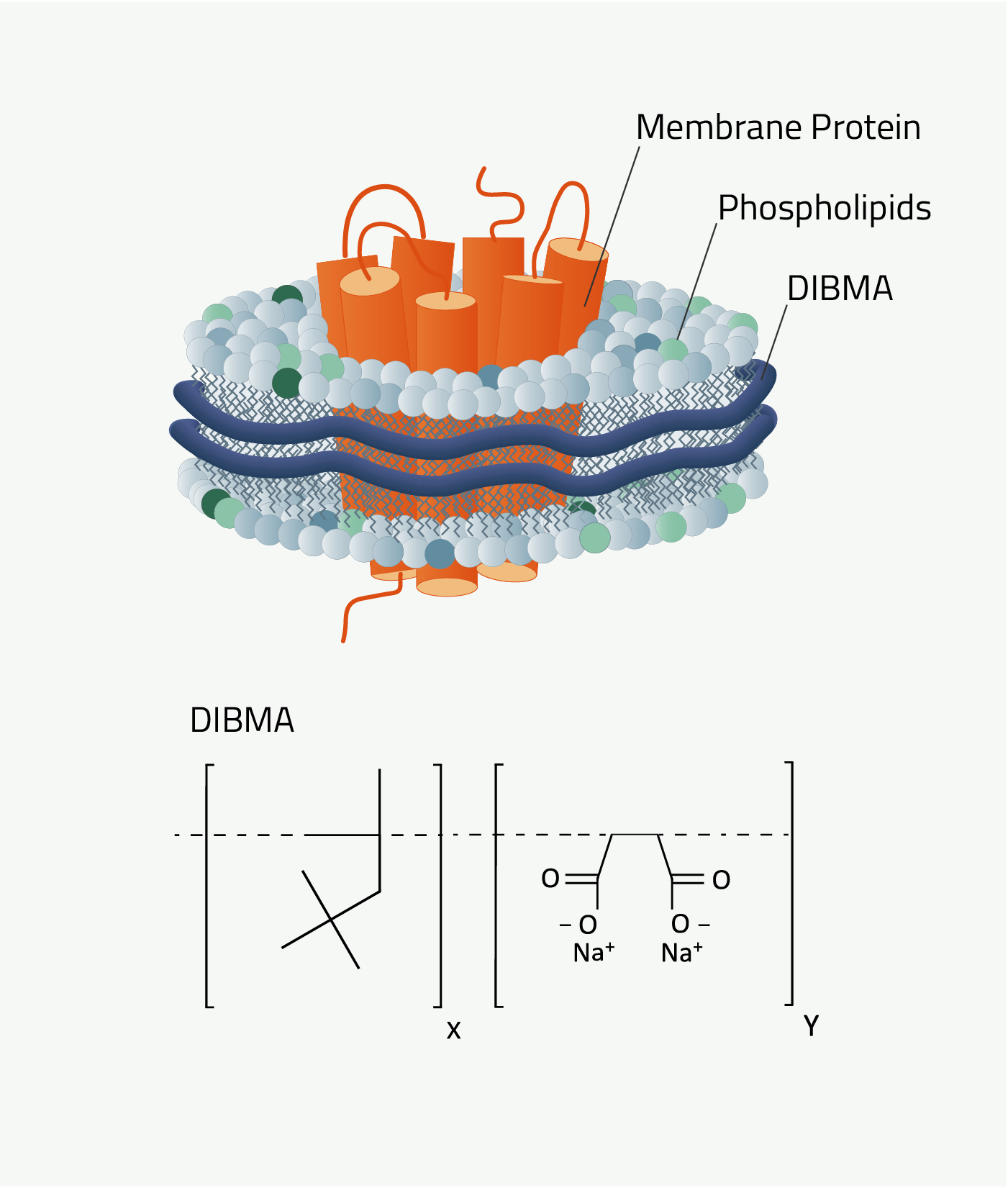
DIBMA
DIBMA is short for Diisobutylene-maleic acid. The Copolymer Nanodisc that it forms are called "DIBMA-Lipid-Particles", DIBMALP for short. It shares the same protocol as all other Copolymers that are featured here.
Why should you choose DIBMA?
DIBMA has one key advantage over SMA and AASTY. As it can be seen by comparing figures 4 and 7 DIBMA is missing the aromatic ring that the other two Copolymers have. And this has great implications.
DIBMA does not absorb light at a wavelength of 280nm compared to SMA and AASTYs. This is crucial since this wavelength is also used for protein quantification via absorbance measurements. SMA and AASTY nanodiscs cannot be used for that as their aromatic ring distorts the measurements for protein quantification.
DIBMA does not absorb light at a wavelength of 280nm compared to SMA and AASTYs. This is crucial since this wavelength is also used for protein quantification via absorbance measurements. SMA and AASTY nanodiscs cannot be used for that as their aromatic ring distorts the measurements for protein quantification.
Why should you not use DIBMA?
The biggest counterargument to DIBMA is that compared to SMA and AASTY its solubilization rates can be lower. Meaning that (on average) DIBMA nanodiscs - also called DIBMALPs - result in lower amounts of stabilized membrane protein of interest than other Copolymers.
However, this is only a rule of thumb. Some proteins did indeed solubilize at a higher rate with DIBMA compared to other Copolymers. Another reason why screening will always remain important.
However, this is only a rule of thumb. Some proteins did indeed solubilize at a higher rate with DIBMA compared to other Copolymers. Another reason why screening will always remain important.
DIBMA compared to detergents
For the longest time, the science behind membrane proteins relied on detergents for both solubilization and stabilization. However, detergents such as DDM or LMNG come with their sets of problems. A time-consuming screening process for the correct detergent and the constant need to add it to all buffers can be avoided. But this applies to all Copolymer Nanodisc.
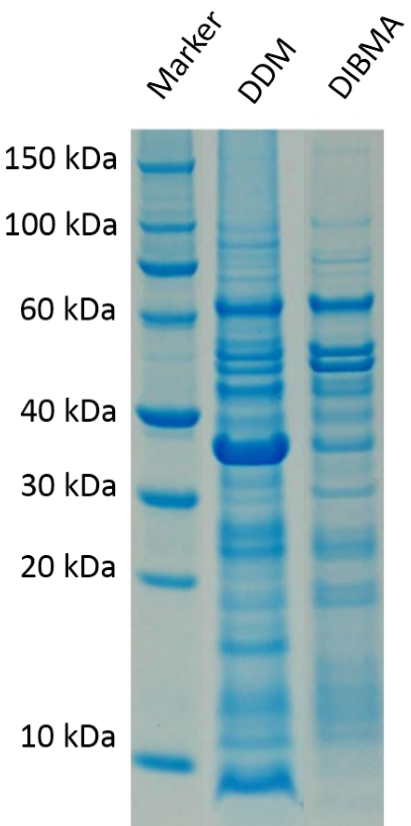
Figure 4 shows the amount of membrane protein of interest was stabilized compared between a DIBMALP and the same construct using the detergent DDM. As it can clearly be seen DDM has way fewer specific bands at the desired kDA values of around 40-60 kDa.
Figure 4 shows the amount of membrane protein of interest was stabilized compared between a DIBMALP and the same construct using the detergent DDM. As it can clearly be seen DDM has way fewer specific bands at the desired kDA values of around 40-60 kDa.
Types of DIBMA
There are different types of DIBMAs that need to be differentiated.
- Different buffered DIBMAS - TRIS or HEPES buffer
- DIBMAs of different lengths - DIBMA 10 and 12 build different large Copolymer chains
- Differently charged DIBMAS - to make DIBMAS more tolerant to divalent cations Glycerol and Glucosamine groups have been attached to the original DIBMA structure
| DIBMA - Features in detail | |
|---|---|
| Usage | Protein solubilization |
| Formula Weight | ~12,000 g/mol or 10,000 g/mol |
| pH | 7.5 in buffer |
| dn/dc | 1.35 M-1 |
| Solubility | > 10 % in H2O |
| Absorbance at 280 nm | < 0.3 (1 % solution) |
| Mg2+ Tolerance | Dependent on DIBMA product Increased with less charged DIBMAs |
| Ca2+ Tolerance | Dependent on DIBMA product Increased with less charged DIBMAs |
A good publication to read up even more details about DIBMA is Oluwole et al. 2017
Frequently asked questions regarding DIBMA
Is DIBMA from Cube Biotech ready to use?
Yes, our DIBMA is ready to use. You can start directly with solubilization. Read our protocol for more information.
Which pH is suitable for DIBMA?
For DIBMA a pH of 7.5 is recommended. DIBMA does not solubilize if the pH is smaller than 6.5.
Which concentrations of DIBMA should I use for my protein?
In general, we advise you to add 2,5 % DIBMA to your solution. But the optimal conditions have to be screened by yourself (Fig. 4).
I used DIBMA for protein solubilization and a white precipitate appeared - what happened?
Your DIBMA precipitated. You should check your pH and ensure that your pH never drops down to 6.5.
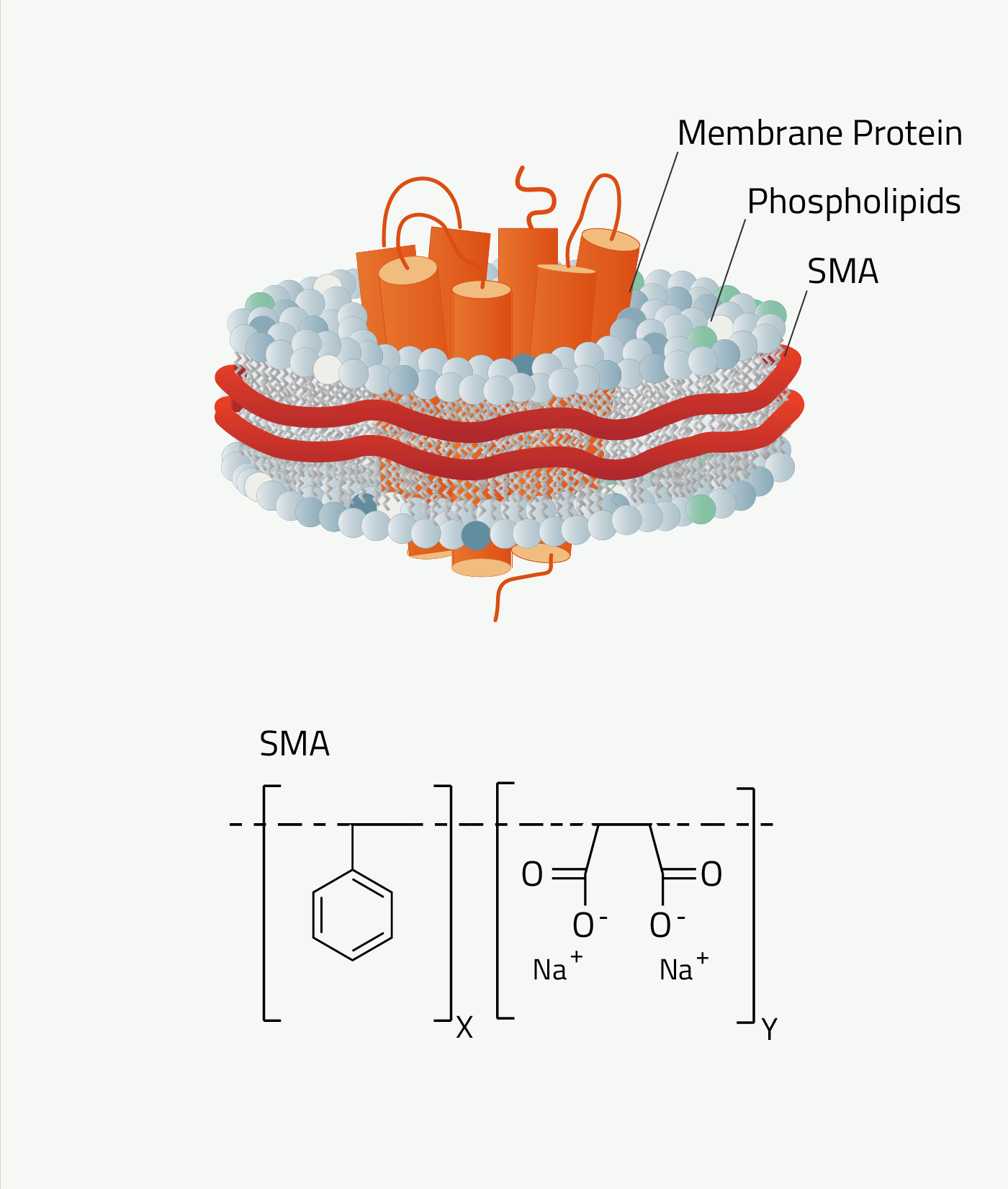
SMA
SMA is short for styrene maleic anhydride. The Copolymer Nanodisc that it forms are called "SMA-Lipid-Particles", SMALP in short.
Why should you choose SMA?
SMA is the Copolymer that has been the longest in use to create Copolymers. Therefore, SMA has the greatest proven database for successfully solubilized membrane proteins. Enough to spawn its own scientific community, the SMALP network.
Compared to DIBMA, SMA has higher solubilization rates of membrane proteins. This means, that one can get more membrane protein with SMALP compared to DIBMALPs.
Compared to DIBMA, SMA has higher solubilization rates of membrane proteins. This means, that one can get more membrane protein with SMALP compared to DIBMALPs.
Why shouldn't you choose SMA?
As can be seen in figure 7 SMA contains an aromatic ring. This results in SMA absorbing light at wavelengths of 280nm. This wavelength is normally used to quantify proteins. So, membrane proteins solubilized by SMALPs cannot be quantified that way, as SMALPs distort the measurement. DIBMA does not have this problem.
| SMA - Features in detail | |
|---|---|
| Usage | Protein solubilization & stabilization |
| Molecular weight of Copolymer molecules | ~5,000 g/mol to 10,000 kDa |
| pH | 7.5 in buffer |
| Divalent cationic tolerance | Usually ~5 mM with one SMALP at 100 mM |
| Absorbance at 280 nm | Yes |
Frequently asked questions regarding SMA
What SMA is the best one to use?
This is a tricky question, as it depends on the membrane protein which SMA performs best, however, SMA 30010S is in general the most recommended. Keep in mind that this is no guarantee for it to be the best but in our experience, it is a good allrounder.
What are the recommended concentrations of SMA?
Mix the supplied solutions at a concentration of 1-5% SMALP to the total solution. These conditions can be used as indications. The optimal SMA concentration must be determined separately for each experiment.
How is SMA stored?
Store in cool and dry places, protected from direct sunlight. Long-term storage is recommended at 4°C
The viscosity of my SMA has decreased, what happened?
This can occur by storing SMA at low temperatures (as recommended!). Simply warm the SMA solution up to room temperature to restore its original viscosity.
How do I quantify the amount of protein after SMA treatment?
In contrast to DIBMA, SMA absorbs light at 280 nm similar to proteins. Therefore ultraviolet absorption is no option here. However other protein quantification assays are viable options like:
- Bicinchoninic Acid (BCA)
- Bradford
- Folin-Lowry in some cases
AASTY
Poly(acrylic acid-co-styrene), AASTY, or SAA, is a highly-alternating copolymer composed of acrylic acid and styrene.
AASTY is well-suited for native lipid nanodiscs and is effective in the extraction of membrane proteins from mammalian, bacterial, and yeast systems (1,2). The use of AASTY for native nanodisc preparations was developed by Dr. Anton Autzen and Dr. Henriette Autzen, in collaboration with the Appel Research Group at Stanford University, and the Yifan Cheng Lab at the University of California San Francisco.
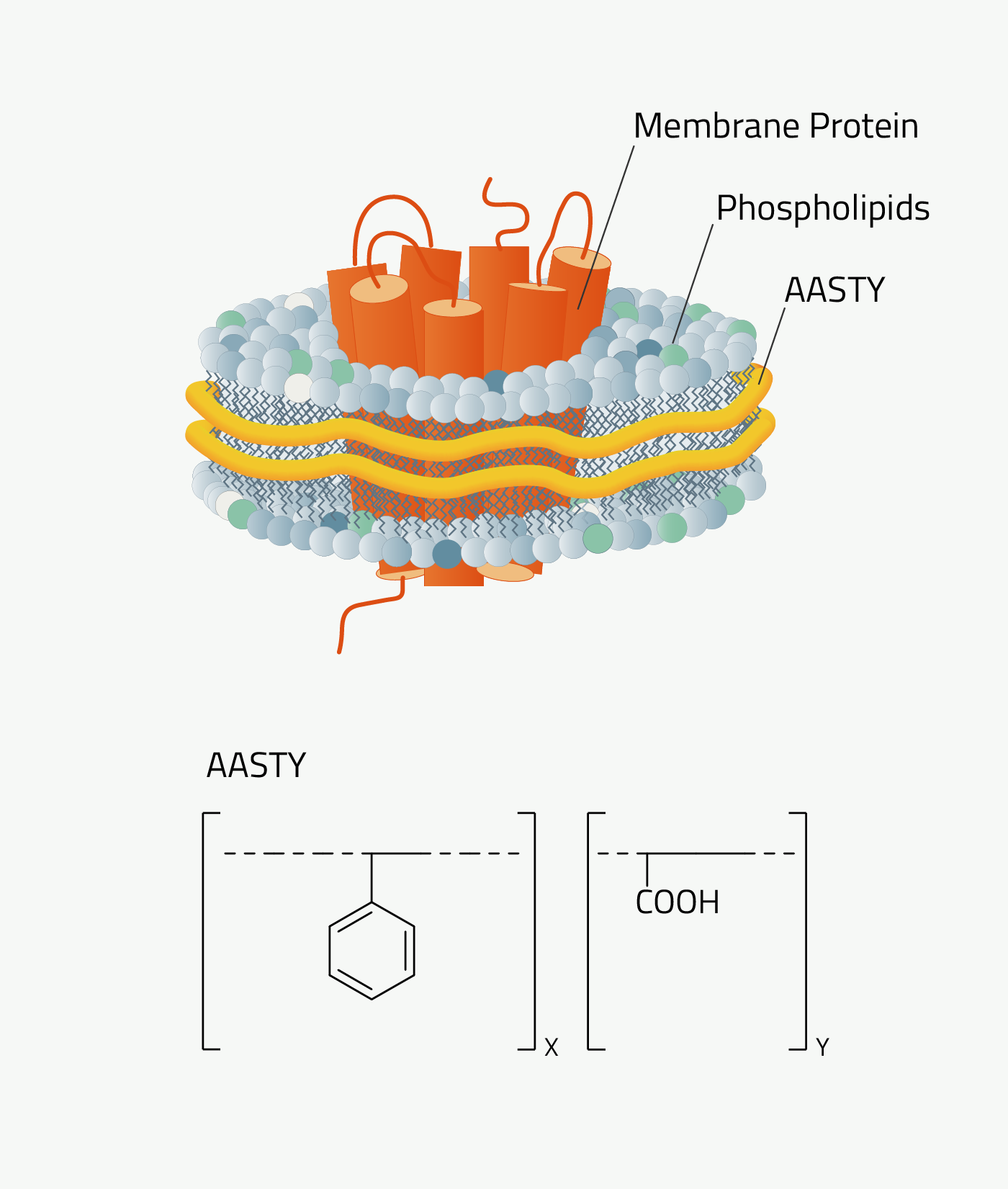
Why should you use AASTY
The reactivity ratios of styrene and acrylic acid allow for a highly alternating monomer sequence in the copolymer (1). This means that they are more homogeneous and presumably more efficient than copolymers that are less homogeneous.
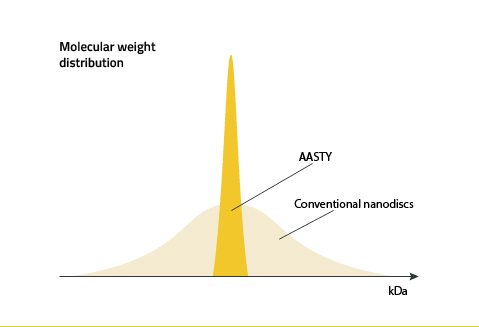
Excess AASTY copolymer can be removed by dialysis after nanodisc formation (3). This is because the dispersity of the Copolymers molecular weight distribution is low and the molecular weights are highly controlled in the reversible addition−fragmentation chain-transfer reaction used to synthesize AASTY. Figure 9 illustrates this.
Excess AASTY copolymer can be removed by dialysis after nanodisc formation (3). This is because the dispersity of the Copolymers molecular weight distribution is low and the molecular weights are highly controlled in the reversible addition−fragmentation chain-transfer reaction used to synthesize AASTY. Figure 9 illustrates this.
Why not use AASTY
One of the major challenges when working with anionic copolymers is their sensitivity to divalent cations: Too high divalent cation concentrations lead to copolymer precipitation and therefore loss of function. As an anionic copolymer, AASTY is sensitive toward divalent cations. This sensitivity depends on the acrylic acid content in the copolymer and can be further mitigated by adding increasing amounts of salt such as KCl and NaCl (3). AASTY copolymers tolerate higher concentrations of Mg2+ than Ca2+, both in nanodiscs and in absence of lipids.
In general, we recommend Timcenko et al. (2022) and Smith et al. (2020) for you to read up on AASTY.
Frequently asked questions regarding AASTY
Is AASTY from Cube Biotech ready to use?
Yes, AASTY is ready to use. You can start directly with solubilization. Read our protocol for more information.
Which pH is suitable for AASTY?
A pH from 7 and up is possible. AASTY does not solubilize if the pH is less than 6.5. As the different AASTY offered differ in composition, they will likely have different optimum pH, dependent on the composition of lipids being solubilized into nanodiscs.
The degree of protonation, and thus the hydrophilic/hydrophobic balance of the copolymer differs across ranges of pH. The effectivity of AASTY for a given protein can be optimized in terms of pH and copolymer used.
The degree of protonation, and thus the hydrophilic/hydrophobic balance of the copolymer differs across ranges of pH. The effectivity of AASTY for a given protein can be optimized in terms of pH and copolymer used.
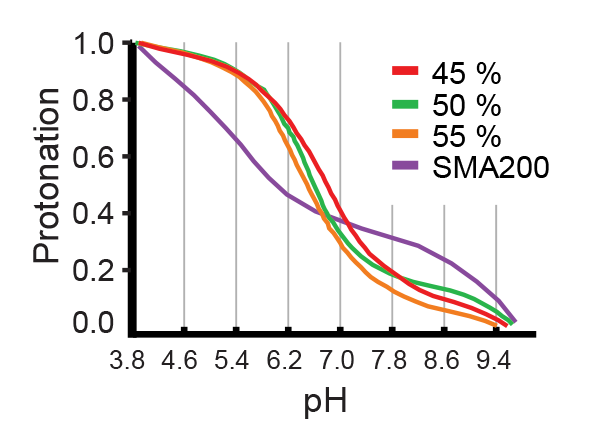
Which concentrations of AASTY should I use for my protein?
You should screen the polymer to membrane protein ratio as this is system-dependent.
I used AASTY for protein solubilization and a white precipitate appeared - what happened?
AASTY likely precipitated. You should check the pH and ensure that it never drops below 7.0.
How do I quantify the amount of protein after AASTY treatment?
AASTY absorbs light at 280 nm similar to proteins due to the styrene moieties. Therefore, UV absorption is no option here. Components in Bicinchoninic Acid (BCA) assays are likely influenced by AASYY. However, the Bradford assay and SDS-PAGE work.
References
- Smith, A. A. A.; Autzen, H. E.; Faust, B.; Mann, J. L.; Muir, B. W.; Howard, S.; Postma, A.; Spakowitz, A. J.; Cheng, Y.; Appel, E. A. Lipid Nanodiscs via Ordered Copolymers. Chem 2020, 6 (10), 2782–2795. https://doi.org/10.1016/j.chempr.2020.08.004.
- Kopf, A. H.; Lijding, O.; Elenbaas, B. O. W.; Koorengevel, M. C.; Dobruchowska, J. M.; van Walree, C. A.; Killian, J. A. Synthesis and Evaluation of a Library of Alternating Amphipathic Copolymers to Solubilize and Study Membrane Proteins. Biomacromolecules 2022. https://doi.org/10.1021/acs.biomac.1c01166.
- Timcenko, M.; Autzen, A. A. A.; Autzen, H. E. Characterization of Divalent Cation Interactions with AASTY Nanodiscs. ACS Appl. Polym. Mater. 2022. https://doi.org/10.1021/acsapm.1c01507
AMPHIPOLS
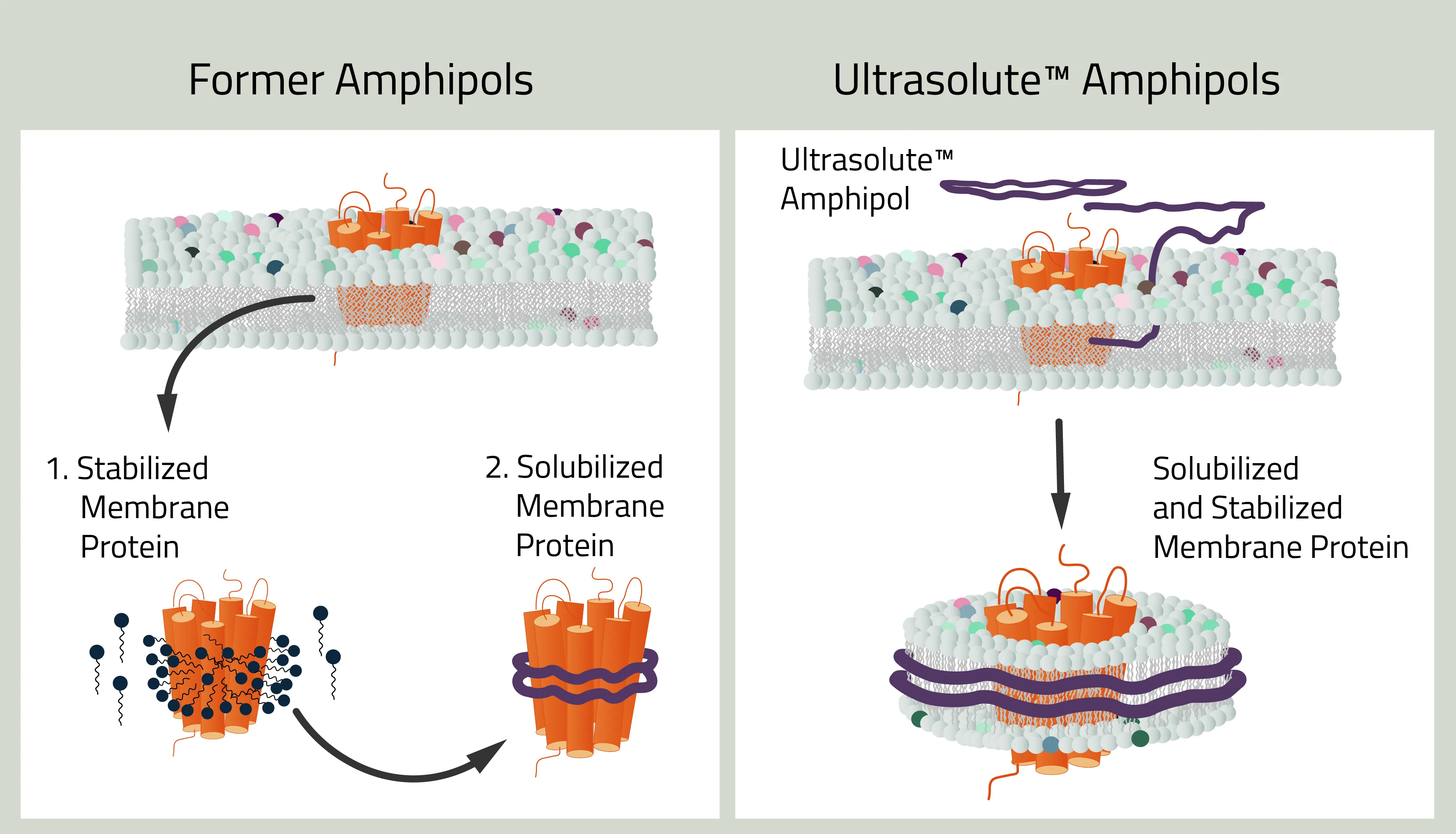
Amphipol is short for "amphiphilic polymers". This type of Copolymer was one of the first developments in this field. However, a lot has changed compared to the "original" amphipols.
Amphipols can be seen as the first major success to use Copolymers for membrane protein stabilization. Their first mention of this use is from Tribet et al. 1996. At that time amphipols were a great alternative to detergents for the stabilization of membrane proteins. But the continuous development of amphipols led to what is called Ultrasolute™ Amphipols.
Amphipol - I heard that name before
As mentioned before, the history of amphipols can be dated back to 1996. Back then amphipols could only stabilize but not solubilize a membrane protein. This first task still needed to be done with detergents, similar to MSP nanodiscs. But recent development changed that. The new Ultrasolute™ Amphipols combine both steps in a fast and easy manner, comparable to our other Copolymers. Other advancements of Amphipols during these 2 decades can be read up in Marconnet et al. 2020 and Marconnet et al. 2022
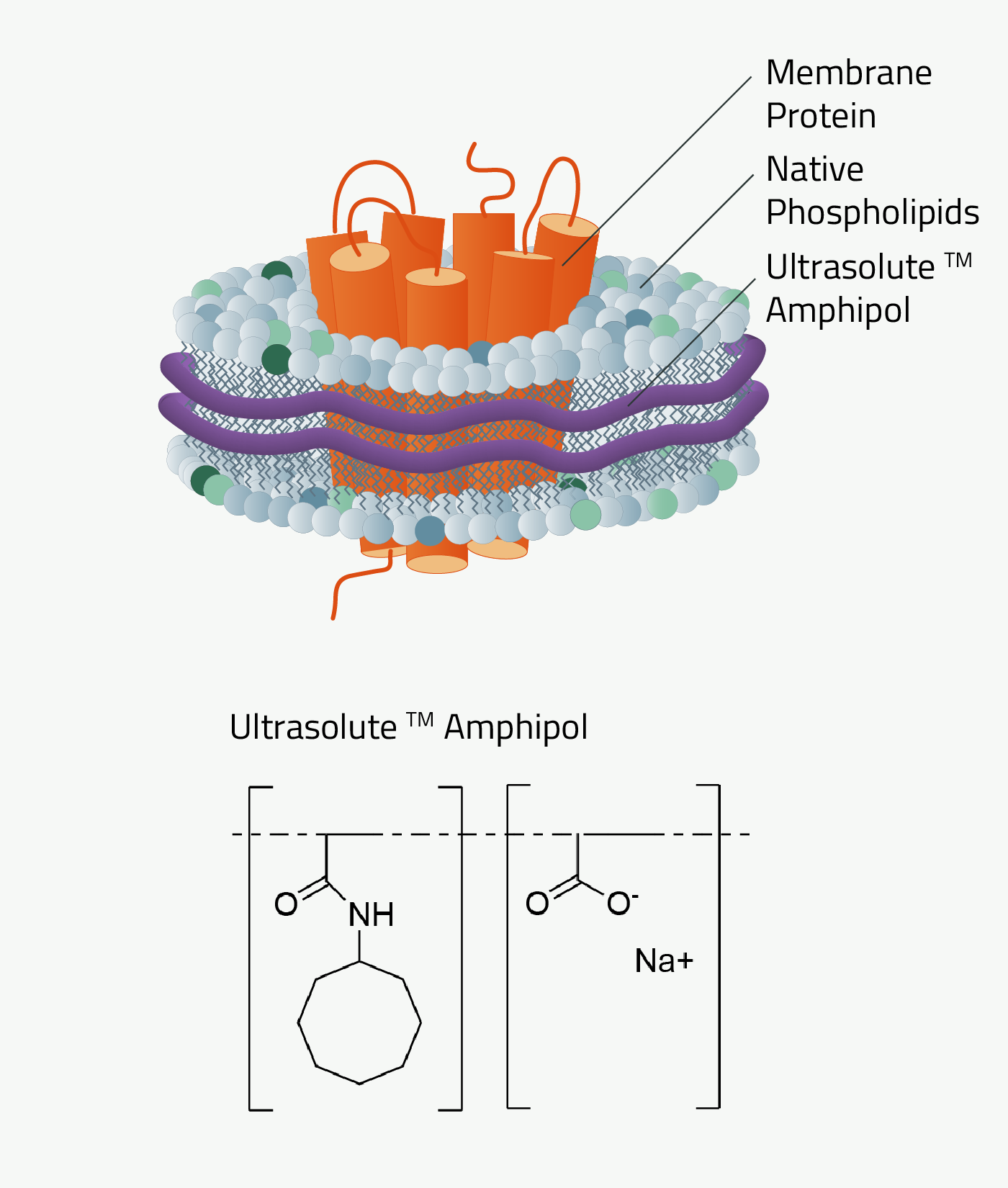
Why should you use Ultrasolute™ Amphipols?
Amphipols combine two of the greatest advantages of other Copolymer Nanodisc polymers. First, their solubilization efficiency of membrane protein is at least on par with SMA. Simultaneously it has no noteworthy absorption at wavelengths 280nm compared to SMA and even less absorption compared to DIBMA. It, therefore, does not interfere with size exclusion chromatography or protein quantification measurements.
| Ultrasolute™ Amphipols. - Features in detail | |
|---|---|
| State | Lyophilized powders to be solved in water |
| Prefered Buffer | HEPES |
| pH after solving of powder | 7.5 |
| Divalent cationic tolerance | at leat 10 mM |
| Absorbance at 280 nm | < 0.1 (280 nm, 1% solution) |
References
- Tribet C. et al. (1996) Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions, PNAS Vol 93; https://doi.org/10.1073/pnas.93.26.15047
- Marconnet A. et al. (2020) Amphipols: Solubilization and Stabilization of Membrane Proteins by Cycloalkane-Modified Amphiphilic Polymers,10.1021/acs.biomac.0c00929
- Marconnet A. et al. (2022) Influence of Hydrophobic Groups Attached to Amphipathic Polymers on the Solubilization of Membrane Proteins along with Their Lipids, doi.org/10.1021/acs.analchem.2c01746


