PureCube Ni-NTA Agarose
Order number: 31103
Description
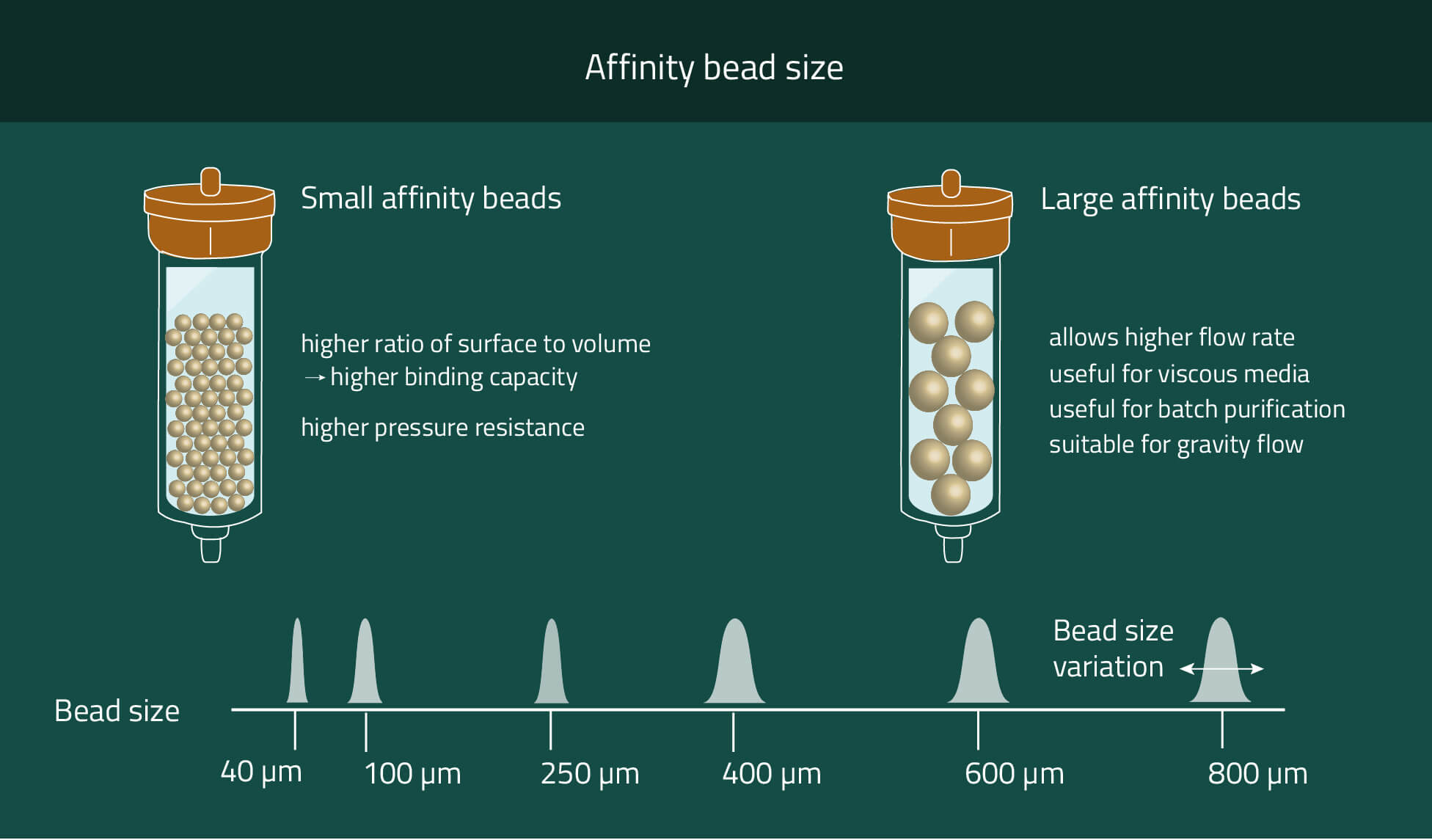
Our PureCube Ni-NTA agarose resins are your best option to purify His-tagged protein using FPLC, Batch spin, or column chromatography. Their size range from market standard 40 µm and 100 µm diameters to 400 µm (XL).
These three options provide plenty of possibilities to choose the correct agarose resin for your purpose. Chose smaller beads for higher mechanical stability and higher protein yield. Meanwhile bigger beads provide faster flow rates and the possibility to work with very viscous cell media.
Furthermore, you can also get these beads pre-packed in a column/cartridge or as magnetic beads.
These three options provide plenty of possibilities to choose the correct agarose resin for your purpose. Chose smaller beads for higher mechanical stability and higher protein yield. Meanwhile bigger beads provide faster flow rates and the possibility to work with very viscous cell media.
Furthermore, you can also get these beads pre-packed in a column/cartridge or as magnetic beads.
Datasheets
- Ni-NTA (40µm) Datasheet
- Ni-NTA (100µm) Datasheet
- Ni-NTA (400µm) Datasheet
- Native Purification Protocol
- Denaturing Purification Protocol
- Batchspin MINI Native Purification Protocol
- Batchspin MINI Denaturing Purification Protocol
- Batchspin MIDI Native Purification Protocol
- Batchspin MIDI Denaturing Purification Protocol
- Washing & Regeneration Protocol
- Cartridge Packing Protocol
| Feature | |
|---|---|
| Usage | Specific binding and purification of 6x His tagged proteins |
| Specificity | Affinity to His tagged proteins |
| Binding capacity | 20 - 80 mg/ml, depending on bead size |
| Bead Ligand | Ni-NTA |
| Bead size | 40 μm, 100 µm, or 400 µm |
| Chelator stability | Stable in buffer containing 10 mM DTT and 1 mM EDTA |
| Filling quantity | Delivered as a 50 % suspension |
| Required equipment |
|
Citations
Lab Results
High yield and purity
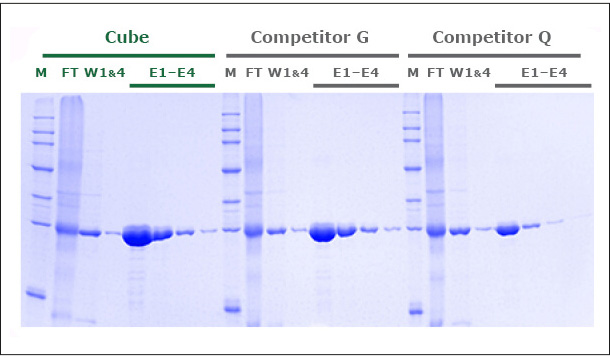
Our unique production process yields a Ni-NTA Agarose that exhibits a protein binding capacity >20% higher than that of two leading competitor products. Figure 1 shows the SDS-PAGE of GFP expressed in E. coli and purified in gravity colums with PureCube Ni-NTA Agarose and the Ni-NTA resin from Competitor G and Competitor Q. The protein yield in 4 elutions (E1-E4, Cube) was 80 mg/mL, compared to 65 and 48 mg/mL obtained with the alternative resins (E1-E4, Competitor G, Competitor Q). Similar results (10-18% higher binding capacity; data not shown here) were obtained comparing the purification of JNK1 (Kinase, 48 kDa) on PureCube Ni-NTA and the Ni-NTA of leading providers.
Our unique production process yields a Ni-NTA Agarose that exhibits a protein binding capacity >20% higher than that of two leading competitor products. Figure 1 shows the SDS-PAGE of GFP expressed in E. coli and purified in gravity colums with PureCube Ni-NTA Agarose and the Ni-NTA resin from Competitor G and Competitor Q. The protein yield in 4 elutions (E1-E4, Cube) was 80 mg/mL, compared to 65 and 48 mg/mL obtained with the alternative resins (E1-E4, Competitor G, Competitor Q). Similar results (10-18% higher binding capacity; data not shown here) were obtained comparing the purification of JNK1 (Kinase, 48 kDa) on PureCube Ni-NTA and the Ni-NTA of leading providers.
Superior DTT and EDTA stability
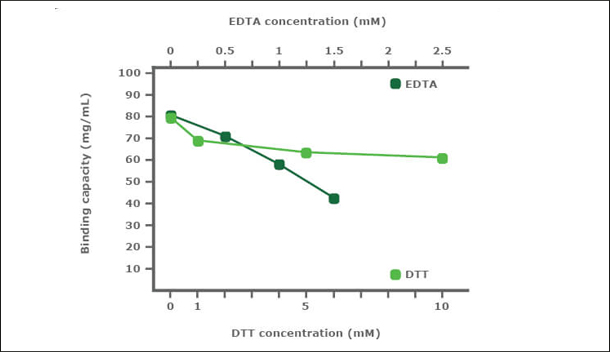
PureCube Ni-NTA Agarose is very robust in the presence of DTT and EDTA. In a stability test, PureCube Ni-NTA Agarose was exposed to increasing concentrations of DTT or EDTA for 1 h. Thereafter, the resins were used to purify E. coli-expressed GFP-His in gravity columns. The binding capacity of the resin decreased in the presence of both DTT and EDTA but the decay rate was shallow. In presence of DTT, PureCube Ni-NTA Agarose lost on average 8% binding capacity with each increase in DTT concentration, resulting in an overall decay of 22% at 10 mM. Even at 1.5 mM EDTA, the resin still exihibits 54% of its maximum binding capacity (Fig. 2).
PureCube Ni-NTA Agarose is very robust in the presence of DTT and EDTA. In a stability test, PureCube Ni-NTA Agarose was exposed to increasing concentrations of DTT or EDTA for 1 h. Thereafter, the resins were used to purify E. coli-expressed GFP-His in gravity columns. The binding capacity of the resin decreased in the presence of both DTT and EDTA but the decay rate was shallow. In presence of DTT, PureCube Ni-NTA Agarose lost on average 8% binding capacity with each increase in DTT concentration, resulting in an overall decay of 22% at 10 mM. Even at 1.5 mM EDTA, the resin still exihibits 54% of its maximum binding capacity (Fig. 2).
Robust against oxidation and regenerable
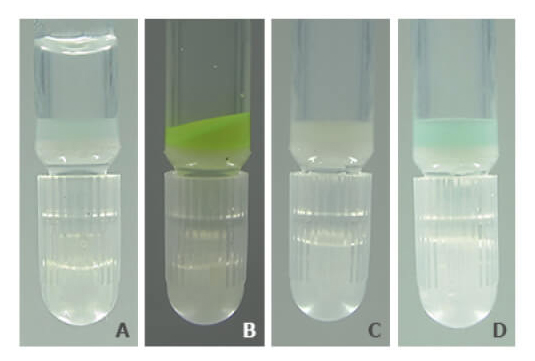
PureCube Ni-NTA Agarose retains its color and function after exposure to as much as 10 mM DTT. Figure 3 shows a photo series of the resin after a 1 h exposure to 5 mM DTT. Unlike other resins, PureCube Ni-NTA Agarose did not turn brown (A). The resin was still able to bind GFP (B), with a measured binding capacity of 65 mg/mL (see Fig. 2). The resin could then be regenerated by stripping the NTA, turning the resin white (C), and reloading it with nickel ions (D). The protocol for regenerating PureCube Ni-NTA Agarose can be downloaded.
PureCube Ni-NTA Agarose retains its color and function after exposure to as much as 10 mM DTT. Figure 3 shows a photo series of the resin after a 1 h exposure to 5 mM DTT. Unlike other resins, PureCube Ni-NTA Agarose did not turn brown (A). The resin was still able to bind GFP (B), with a measured binding capacity of 65 mg/mL (see Fig. 2). The resin could then be regenerated by stripping the NTA, turning the resin white (C), and reloading it with nickel ions (D). The protocol for regenerating PureCube Ni-NTA Agarose can be downloaded.
Video
Video Guide - How to pack FPLC cartridges
Video Guide - FPLC
Video Guide - Column Chromatography
Video Guide - Batch Spin Chromatography
FAQ
Can I get the datasheets for the Ni-NTA resins?
Yes. Just chose the datasheet you need from the list below.
What are the reasons for nonspecific binding?
Some other histidine-rich proteins can also bind to nickel. But washing with NaOH after elution of your protein of interest removes unspecific bound proteins from your resin.
I want to use a high concentration of EDTA and DTT. Is it possible to use Ni-NTA from Cube Biotech?
No, it is not recommended because nickel is reduced with DTT or dissolved with EDTA. If you want to use high concentrations of EDTA and DTT you should use our INDIGO-Ni resin.
How is the capacity at high flow rates?
If higher flow rates are desired we recommend using beads with bigger diameters. We offer Ni-NTA beads with mean diameters of 40µm, 100µm, and 400µm (XL).
With each size increase, the flow rates also increase due to the proportionally increasing space between the beads. However, the surface of the beads does not increase at the same speed as the diameter (square-cube-law). That results in decreasing amounts of purified protein per mL beads while increasing the bead sizes.
For 40µm of 100µm beads, we both have average purification amounts of ~80 µg protein/mL beads. With 400 µm (XL) beads, this decreases to ~20 µg/mL.
We recommend reading the corresponding section of the "Introduction to agarose matrixes" guide on this subject for more detailed information.
With each size increase, the flow rates also increase due to the proportionally increasing space between the beads. However, the surface of the beads does not increase at the same speed as the diameter (square-cube-law). That results in decreasing amounts of purified protein per mL beads while increasing the bead sizes.
For 40µm of 100µm beads, we both have average purification amounts of ~80 µg protein/mL beads. With 400 µm (XL) beads, this decreases to ~20 µg/mL.
We recommend reading the corresponding section of the "Introduction to agarose matrixes" guide on this subject for more detailed information.
After using DTT my resin turned orange. How to regenerate it?
The DTT has probably destroyed your beads. Ni-NTA beads only have a limited DTT tolerance of about 1 mM. However, you can regenerate them to regain their functionality. Please read our detailed protocol for more information regarding this.
However, we would recommend using Ni-INDIGO products instead. They work with the same buffers and protocols as the Ni-NTA products but have a DTT tolerance of 20 mM.
However, we would recommend using Ni-INDIGO products instead. They work with the same buffers and protocols as the Ni-NTA products but have a DTT tolerance of 20 mM.