NTA Versus IDA: What's The Difference?
If you are using immobilized-metal affinity chromatography (IMAC) to purify proteins, there is a good chance your metal ion is coupled to the resin matrix via either nitrilotriacetic acid (NTA) or iminodiacetic acid (IDA). There are other chelating ligands used for IMAC resins, but NTA and IDA are the popular ones. Which chelating ligand do you use and why? Does it make a difference? To help our customers make an informed purchasing decision, we took our PureCube Ni-NTA Agarose and PureCube Ni-IDA Agarose and put them under the magnifying glass (figuratively, of course).
This page answers the following questions about NTA and IDA:
- How do the two ligands differ structurally and chemically?
- How does a chelating ligand impact binding capacity?
- What role does the metal ion play in resin affinity and specificity?
- How do NTA and IDA respond to oxidizing agents?
- Does a chelator in the sample buffer impact NTA and IDA differently?
NTA vs. IDA: a Tale of two Ligands
Immobilized metal affinity chromatography (IMAC) is a popular method for protein purification, particularly for recombinant proteins fused to a Polyhistidine-tag. Transition metal ions immobilized to a matrix through a chelating ligand interact with the Polyhistidine-tag, effectively sequestering the fused protein from a sample. Nitrilotriacetic acid (NTA) and iminodiacetic acid (IDA) are two such ligands commonly used in commercially available resins. In this application note, we summarize our knowledge, from publications and in-house experiments, regarding the differences, advantages, and disadvantages of the two ligands. We present a few basic principles that should underly the purchase of a resin to optimize purification results.
Using metals to capture proteins
Immobilized metal affinity chromatography dominates generic single-step protein purification
Since its inception in 1975(1), IMAC, or immobilized metal affinity chromatography with its variations and improvements, has been widely used to purify proteins. In its most popular form, a transition metal ion fixed to a matrix via a ligand interacts with the imidazole rings of a Polyhistidine-tag (a chain of 3 to 10 histidines) fused to the C- or N-terminus of a peptide. The interaction effectively „sequesters“ the protein from the solution, and the bound protein is then dissociated from the metal ion by elution with an imidazole gradient, changes in pH, or metal chelation(2).
Initially developed for native proteins with an intrinsic affinity to metal ions, the application of IMAC has expanded to include phosphorylated proteins, antibodies, and a long list of proteins outfitted with a Polyhistidine-tag. Especially advantageous is the small size and low immunogenicity of the Polyhistidine-tag, which generally does not interfere with protein function(2). Trivalent and tetravalent ions are commonly used for the purification of phosphoproteins, while divalent ions such as Ni2+, Cu2+, and Zn2+ are used for recombinant His-tagged proteins. Low cost and functional simplicity account for much of its popularity but IMAC is also very robust, making this purification system ideal for methods that call for protein-specific conditions. For example, the imidazole-metal ion interaction can take place under native and denaturing conditions, as well as under oxidizing or reducing conditions. Furthermore, IMAC is compatible with a broad spectrum of chemicals(1). Finally, the high affinity, specificity, and scalability of the system enable very efficient purification of proteins even from crude lysates(1).
Initially developed for native proteins with an intrinsic affinity to metal ions, the application of IMAC has expanded to include phosphorylated proteins, antibodies, and a long list of proteins outfitted with a Polyhistidine-tag. Especially advantageous is the small size and low immunogenicity of the Polyhistidine-tag, which generally does not interfere with protein function(2). Trivalent and tetravalent ions are commonly used for the purification of phosphoproteins, while divalent ions such as Ni2+, Cu2+, and Zn2+ are used for recombinant His-tagged proteins. Low cost and functional simplicity account for much of its popularity but IMAC is also very robust, making this purification system ideal for methods that call for protein-specific conditions. For example, the imidazole-metal ion interaction can take place under native and denaturing conditions, as well as under oxidizing or reducing conditions. Furthermore, IMAC is compatible with a broad spectrum of chemicals(1). Finally, the high affinity, specificity, and scalability of the system enable very efficient purification of proteins even from crude lysates(1).
More than protein purification
IMAC is a versatile technology with surprising applications. Immobilizing His-tagged antigens to surfaces enhances the sensitivity of ELISA-based diagnostic tools, and protein-protein interaction studies on functionalized surfaces can be mediated by IMAC. In proteomic studies, IMAC is used for pre-separation steps to reduce the complexity of study samples. Finally, in the Chelex method, samples are incubated with a matrix coupled to an unloaded chelating ligand. The ligand depletes metal ion inhibitors of PCR thereby quickly generating a sample ready for amplification.
Choosing a chelating Ligand: which Tale to use?
The coordination number of the ligand has an effect on the yield and purity of the eluted protein
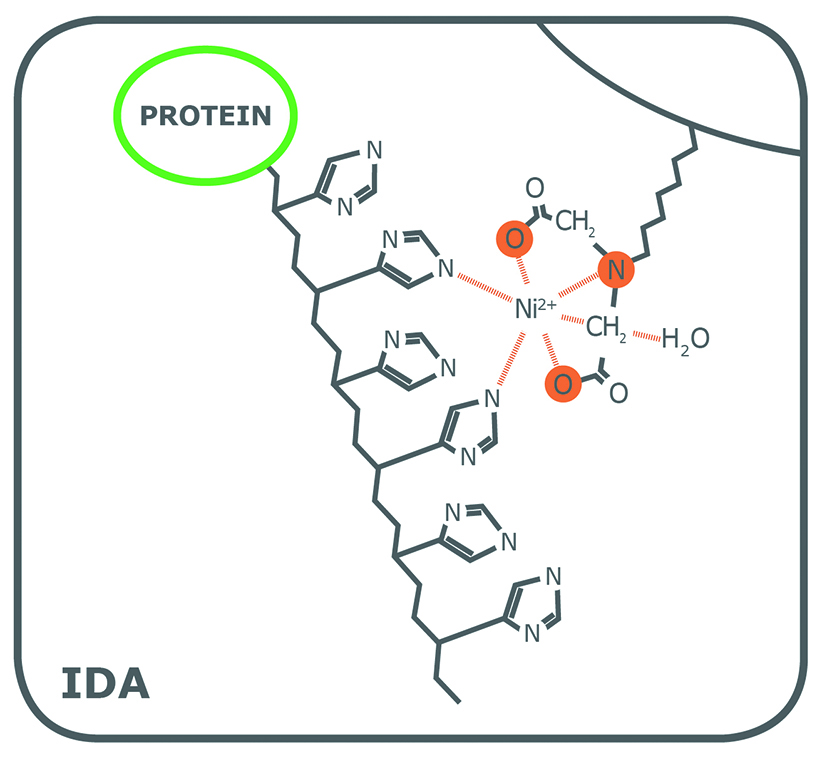
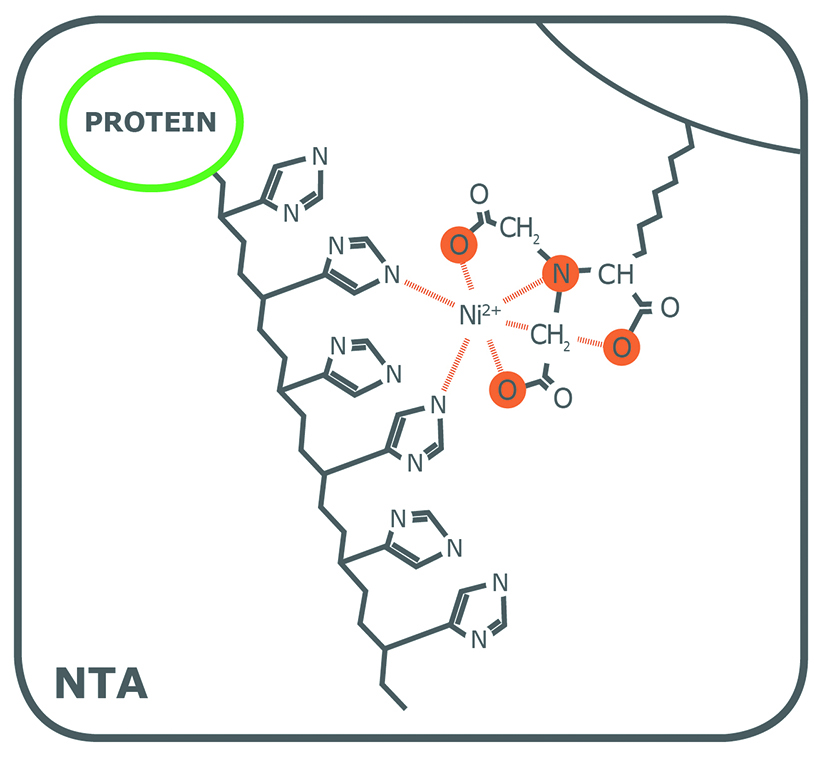
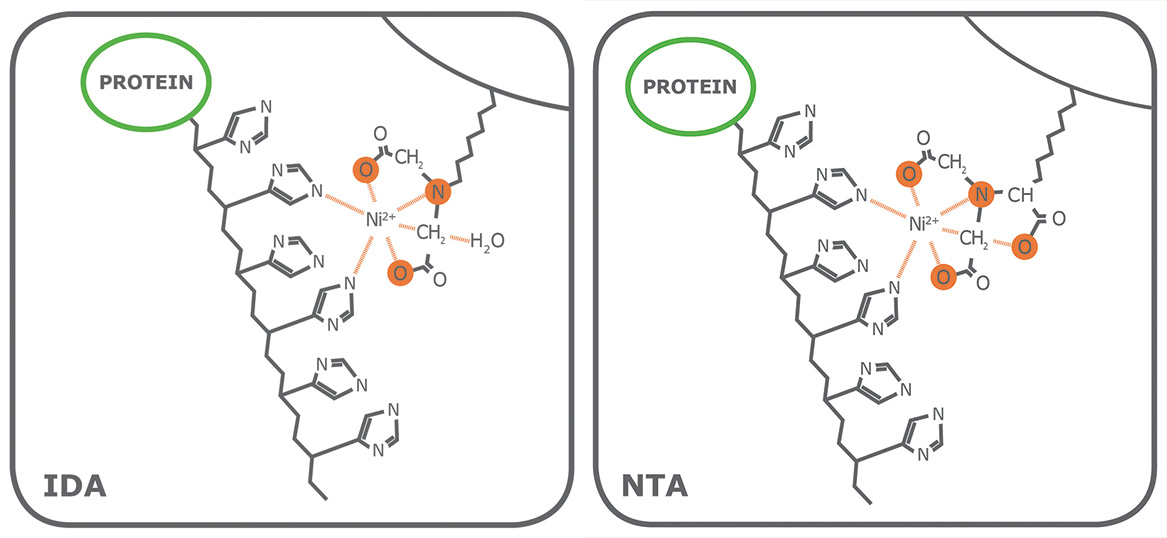
Porath and colleagues(3) developed the first embodiment of an IMAC resin by fixing metal ions to agarose with iminodiacetic acid (IDA). This ligand is still widely used in commercial IMAC resins, along with nitrilotriacetic acid (NTA), first introduced in 1987 by Hochuli and colleagues(4). Since then, other proprietary chelating ligands have been developed (e.g., TED, TALON®), but IDA and NTA represent the most commonly used IMAC resins for affinity purification of proteins. Structurally, IDA and NTA differ by the presence of an additional carboxymethyl group on NTA. Chemically, the additional functional group makes NTA a stronger coordinator of metal ions because it has four valencies available compared to the three of IDA (Fig. 1).
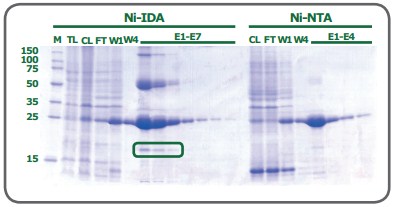
While both ligands have 2 valencies available to interact with the imidazole rings of histidine residues, the coordination number plays a role in the quality of the resulting purified fraction. To begin, as a trivalent ligand (coordination number 3), IDA exhibits higher leaching of metal ions, particularly in the equilibration and wash steps(1). Elution fractions from NTA-based resins also contain leached metal ions, but substantially less than observed from IDA(1). Secondly, while some variation is attributable to the nature of the protein being processed, a His-tagged protein eluted from an IDA resin frequently has lower purity compared to that obtained from an NTA resin. Figure 2 shows the SDS-PAGE of eGFP expressed in E. coli cells and purified on PureCube Ni-IDA and PureCube Ni-NTA resins. The first three elution fractions from the IDA resin show distinct bands for lysate protein components other than the target.
| Ligand | IL-1β-His | JNK1-His | GFP-His |
|---|---|---|---|
| Ni-IDA | 21 mg/ml | 100 mg/ml | 100 mg/ml |
| Ni-NTA | 22 mg/ml | 61 mg/ml | 80 mg/ml |
| Co-NTA | 15 mg/ml | 30 mg/ml | 19 mg/ml |
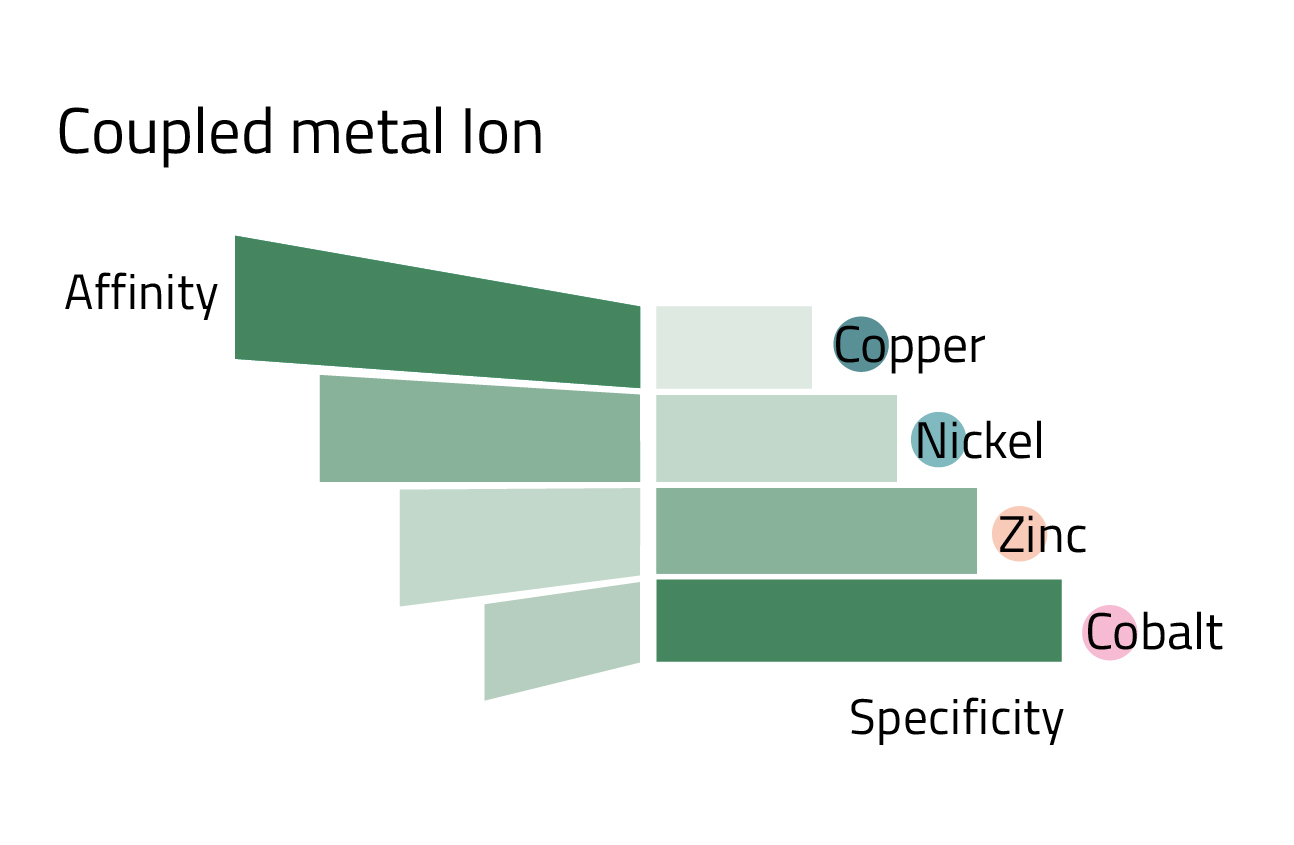
However, IDA has also advantages. Resins constructed with this ligand are generally less expensive. Additionally, the tridentate ligand requires a lower imidazole concentration to elute protein than tetradentate NTA. Lastly, IDA is a smaller molecule that can be coupled to the matrix at a higher density resulting in a higher metal loading capacity. Generally, low loading density enhances the quality of the purification but results in lower target protein recovery, whereas high loading density generates greater yields but with higher unspecific binding. By combining the loading density of IDA or NTA with the affinity and specificity of different metal ions (Fig. 3), it is possible to optimize the purity and yield of purification for a particular protein.
NTA
- Tetravalent, coordination number 4
- Higher binding specificity
- Generally, lower yield
- Stronger coordination of metal ions
- Lower metal ion leaching
IDA
- Trivalent, coordination number 3
- Generally, more unspecific binding
- Increased metal ion leaching
- Higher metal loading density
- Requires lower eluent imidazole concentration
Let your protein determine your ligand and metal ion
When asked, most researchers cannot explain why they use one ligand over another for their His-tagged protein purification. However, it may be advantageous to explore different ligands because the binding capacity of a given IMAC resin is very dependent on the metal ion used and the nature of the protein being purified. A more optimized purification might be achieved with an alternate ligand metal combination. For example, typically IDA has a higher metal ion capacity than NTA (>25 μmol Ni2+ or Cu2+ per mL resin for PureCube IDA Agarose and >15 μmol/mL for PureCube NTA Agarose). Theoretically, IDA avails more binding sites for a target protein resulting in a greater capacity, as apparent in Table 1 with JNK1 and GFP. However, binding capacity varies from one protein to another and can be significantly lower, such as with interleukin-1β. A further consideration is that coupling different metal ions to a resin leads to different affinities for a given His-tagged protein. Cobalt exhibits high binding specificity but low affinity (Fig. 3), and thus we see decreased yields with cobalt-based resins (Table 1). Nickel trades off specificity for higher affinity, which accounts for the observed higher recoveries. One can optimize the purification of a His-tagged protein by exploring different metal ion and ligand combinations. It is important to bear in mind, however, that the described trends do not hold across the board for all resins or all proteins.
What else is in your sample? It can affect the bottom line
The presence of specific compounds in study samples impacts the binding capacity of both IDA and NTA-based resins. DTT is a reducing agent used to stabilize proteins with free sulfhydryl groups and disrupt disulfide bonds in preparation for SDS-PAGE. DTT reduces the metal ions of an IMAC resin, often changing the resin color to brown. In a comparative study, PureCube Ni-IDA Agarose and PureCube Ni-NTA Agarose were exposed to different concentrations of DTT. The binding capacity of both resins dropped non-linearly, but Ni-NTA showed a shallower average decay rate of 8.0% with each increase in DTT concentration, compared to 11.1% for Ni-IDA. The overall decrease in binding capacity at 10 mM DTT was 22% and 30% for Ni-NTA and Ni-IDA, respectively (Fig. 4A).
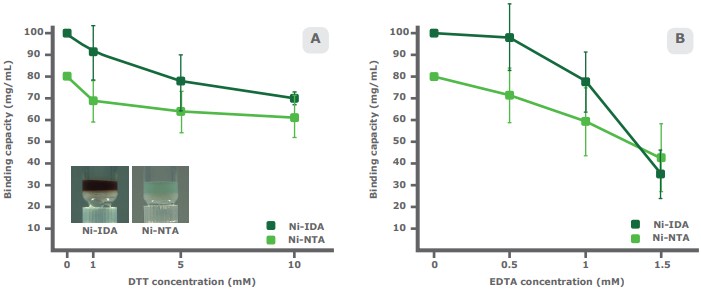
EDTA has a more dramatic impact on binding capacity. This hexadentate chelating agent is found in many buffers and sequesters metal ions. Our comparative study shows that Ni-NTA is more robust than Ni-IDA at higher concentrations of EDTA. Ni-NTA had a non-linear overall drop in binding capacity of 46%, with slightly steeper decay after 1 mM EDTA. Up to 1 mM EDTA, the binding capacity of Ni-IDA decays similar to that of Ni-NTA. Thereafter, the binding capacity drops dramatically (overall decrease: 65%; Fig. 4B).
NTA vs. IDA: making the right Choice
The decision on an IMAC resin for your protein purification needs to consider the following questions:
- How pure does your target protein need to be?
- What yield of target protein do you need?
- Does your sample contain reagents like DTT or EDTA?
- What are your budgetary constraints?
- What yields do you anticipate from your expression?
If you seek high yields and the purity of the recovered protein is of lesser priority, an IDA-based resin might be the right choice. It is less expensive and is still a robust resin that is easily regenerated and reloaded. To minimize unspecific binding, the use of alternate metal ions such as zink and cobalt could be explored. For applications where the purity of the recovered protein is essential (e.g., for subsequent crystallization), NTA-based resins are the most optimal choice. Specificity can be further improved by loading the resin with a metal ion that exhibits highly specific binding (e.g., cobalt). Another advantage of NTA-based resins is their resilience when exposed to agents such as DTT and EDTA, which accommodates a wider range of sample buffers. Resin format is also a deciding factor. Matrix and purification processes can impact final results. Additionally, using excessive resin for a given protein yield can lead to impurities, as binding sites are left exposed.
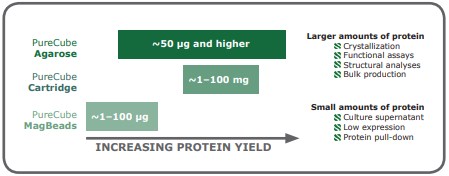
When working with small amounts of protein, it is recommendable to use magnetic beads as a matrix; these are particularly effective for dilute samples, proteins expressed at low levels, and pull-down experiments. Protein amounts standardly obtained from mid-scale suspension culture (>50 μg) are better processed with agarose in batch or gravity column purification. Agarose-based purifications can also be scaled up to bulk production amounts. Finally, packed chromatography columns enable automated processing of protein yields ranging from 1 to 100 mg (Fig 5). In the end, optimizing the purification procedure for a His-tagged protein may require testing a couple of different affinity resin options, but with a few principles, the examination of available IMAC resins can be a systematic process.
Principles of this tale:
- IDA-based resins tend to be less expensive
- NTA-based resins typically generate purer eluates
- Metal ion leaching is significantly lower with NTA-based resins
- Alternate metal ions can adjust the binding specificity of a resin. Co2+ is most specific, then Ni2+, and Zn2+, followed by Cu2+
- NTA-based resins are robust against reducing and chelating agents
Literature
- Block, H., et al. 2009. Immobilized-metal affinity chromatography (IMAC): a review. In: Richard, B.R. and Deutscher, M.P., eds. Methods Enymol 463: 439–473.
- Young, C.L., Britton, Z.T., and Robinson, A.S. 2012. Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol J 7: 620–634.
- Porath, J., et al. 1975. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258: 598–599.
- Hochuli, E., Döbeli, H., and Schacher, A. 1987. New metal chelate adsorbent for proteins and peptides containing neighbouring histidine residues. J Chromatogr 411: 177–184.


