Affinity Chromatography
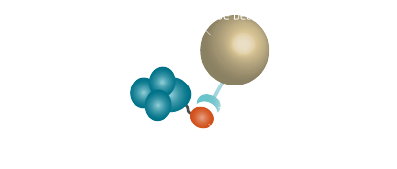
Purification via affinity chromatography is a powerful technique to separate your protein of interest from your sample. The proteins are marked with "tags" (Fig. 3, Table 4) which give them specific binding properties. A suitable immobilized ligand binds to the tagged protein (Fig. 1). So your protein of interest is separated from the rest of your sample.
Principles of Affinity Chromatography
In general affinity chromatography is composed of a stationary phase (solid phase) and a mobile phase (Fig. 1). The mobile phase is your cell lysate or any mixture that contains biomolecules. A ligand that binds the target molecule is attached covalently to the solid phase. The interaction between the solid and the mobile phase are exploited by affinity chromatography to get your desired substance in a pure form. The target molecule binds to the ligand, whereas most of the other molecules flow through. The target biomolecule is eluted by changing conditions (pH or salt concentrations) or by competition with a free ligand.The most important property which the solid phase should have is ligand immobilization. Various materials like acrylates or silica gels are appropriate. To prevent steric interference of the target molecule to the ligand, an inhibitor is attached to the solid phase. This inhibitor is known as the spacer (Fig. 2). The classic spacer is an inhibitor containing a hydrocarbon chain (CH2 spacer). Chemicals like cyanogen bromide or epoxide can functionalize the solid phase with hydrocarbon chains which result in various carbon chain lengths depending on the chemical (Table 2).
The stationary Phase
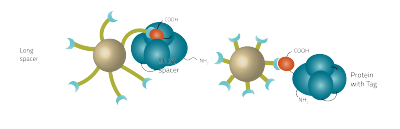
The stationary phase of an affinity chromatography, also called the solid phase, consists of the Core, the Spacer and the Ligand and on occasion a metal ion that is coupled to the ligand (e.g. His-tag purification).
The spacer
The spacer of an affinity bead can vary in size and length. Longer chain lengths have a higher mobility and are thus crucial for stericaly hindered tags.
| Chemical | Chain |
|---|---|
| Cyanogen bromide | C1 |
| Epoxide | C3 |
| Epoxide with C6 acid | C10 |
| Diamin | C10 |
The core
| Bead material | Features |
|---|---|
| Agarose | hydrophilic almost no unspecific bonds the gold standard for protein purification |
| Silica Gel | nanoporous (leads to unspecific bonds) functionalized via silane silanes are washed away by alkaline buffer reduced stability applications: bound nucleic acids chaotropic |
| Aluminium oxide | acidic surface binds amines irreversibly used to reduce the amount of specific substances |
| Acrylate | partially hydrophobic (unspecific binding possible) monodisperse particles used for cell separation |
| Organic polymers | partially hydrophobic (unspecific bounds possible) monodisperse particles can be used for ligand coupling not recommended for protein purification because of unspecific bounds |
Advantages of porous agarose
- easily dispensed to fill and "pack" columns with resin beds of any size
- large enough that biomolecules (proteins, etc.) can flow as freely into and through the beads
- Ligands are covalently attached to the bead polymer (external and internal surfaces) by various means
- Beaded agarose is good for routine applications as it allows a setup, making it suitable for gravity-flow, low-speed-centrifugation, and low-pressure procedures
The ligand
The ligand is responsible for the affinity of the bead. In some cases the affinity of the ligand can be further modified. A noteworthy example here are His-tag purifications. In this case the ligand is a chelator coupled with a metal ion. The metal ion is responsible for the balance between affinity and specificity of the purification.
| Ligand | Target |
|---|---|
| Antibody | Antigen |
| Iron-, aluminium ions | Phosphoproteins |
| Avidin | Biotin |
| Glutathione | His-tagged proteins |
The mobile/liquid phase
Affinity chromatography relies on the presence of an affinity tag - typically a peptide or protein sequence which can be added to the protein of interest on the DNA level. For small tags, this can be done by a two-step PCR, but in most cases dedicated cloning vectors are available. Once equipped with a suitable affinity tag, the gene of interest is expressed and the recombinant protein carries the respective additional amino acids.
Affinity chromatography resins or matrices are typically agarose or magnetic agarose beads that are covalently coupled to a molecule that specifically binds to the affinity tag. There is a great variety in tag-resin partner chemistries and interaction types. See table 3 for a first overview.
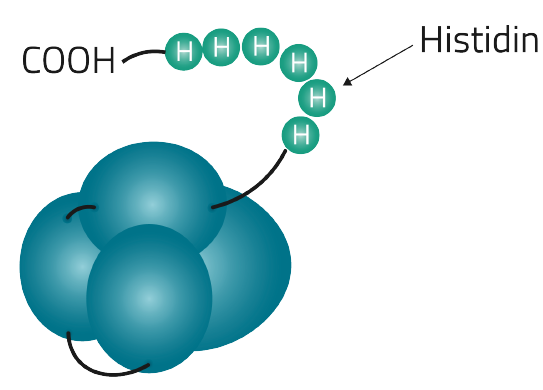
Table 4 on the other hand lists the most commonly used affinity tags and their individual features
| Tag | Amino acid sequence | DNA Sequence | Size | Specificity of interaction (KD) | Protein Yield per ml resin | Elution conditions |
|---|---|---|---|---|---|---|
| Poly His-tag | 6-14 Histidine residues | CAT CAC CAT CAC CAT CAC | 840.8-1,937.9 Dalton | 10 µM | up to 80 mg/ml (1) | Imidazole or histidine, or at low pH |
| GST-tag | GST-tag protein (whole protein) | Plasmid / Basic cloning vector | 26 kDa protein | 1 µM | 10-12 mg/ml | Reduced glutathione |
| Strep®-tag | WSHPQFEK | TGG TCG CAT CCG CAG TTC GAG AAG | 1,058.1 Dalton | 300 nM | up to 9 mg/ml | Desthiobiotin or biotin |
| Rho1D4 / 1D4-tag | TETSQVAPA | ACC GAG ACT TCC CAG GTG GCG CCA GCT | 902.9 Dalton | 20 nM (4) | 3-4 mg/ml | Rho1D4 peptide, low pH, or protease digest |
| FLAG®-tag | DYKDDDDK | GAC TAC AAA GAC GAT GAC GAC AAG | 1,012.9 Dalton | 100 nM | 0.6-1 mg/ml | DYKDDDDK peptide |
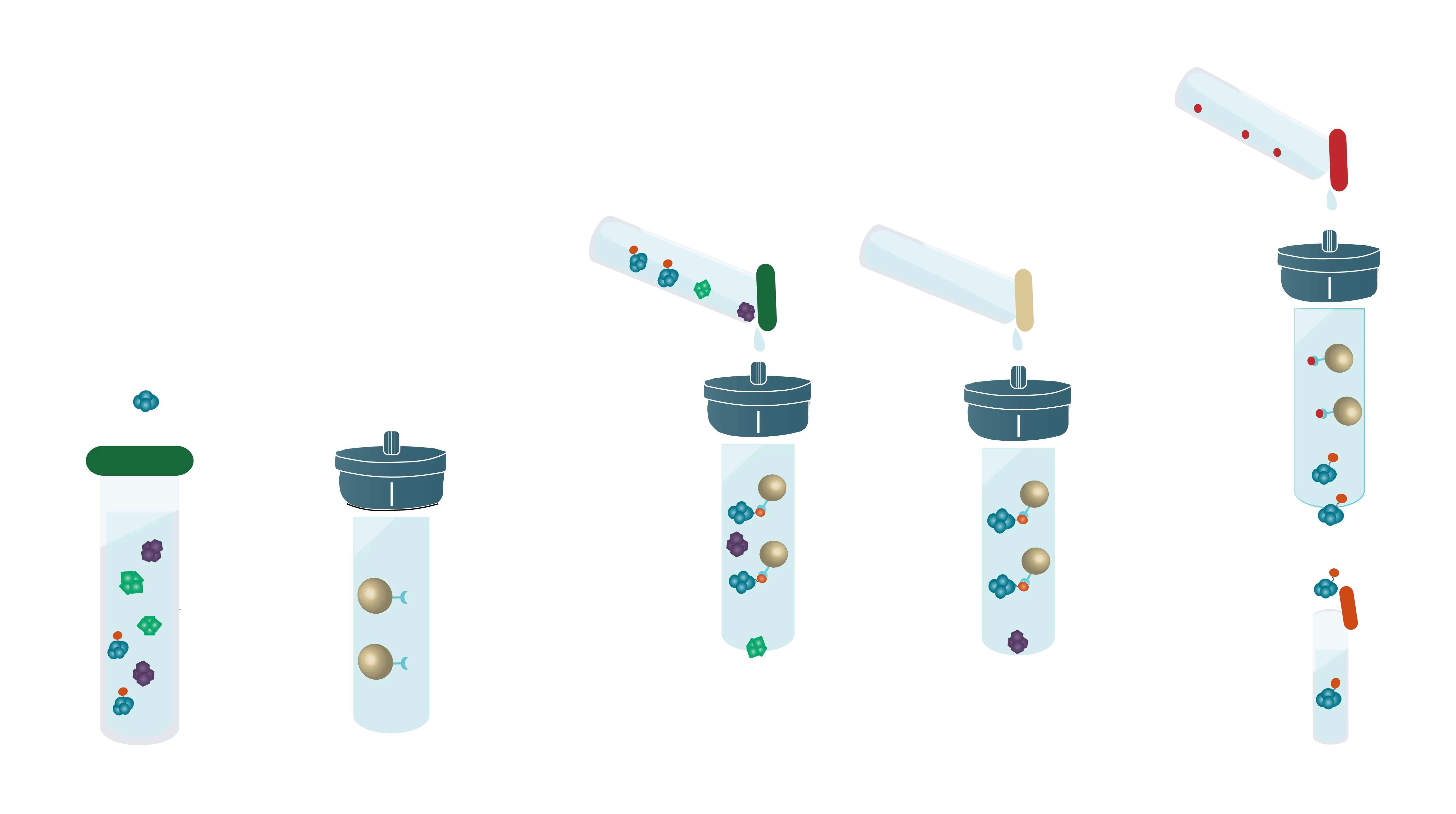
Bind-Wash-Elute: The three steps of Affinity Purification
Most affinity purification protocols follow the same three steps:
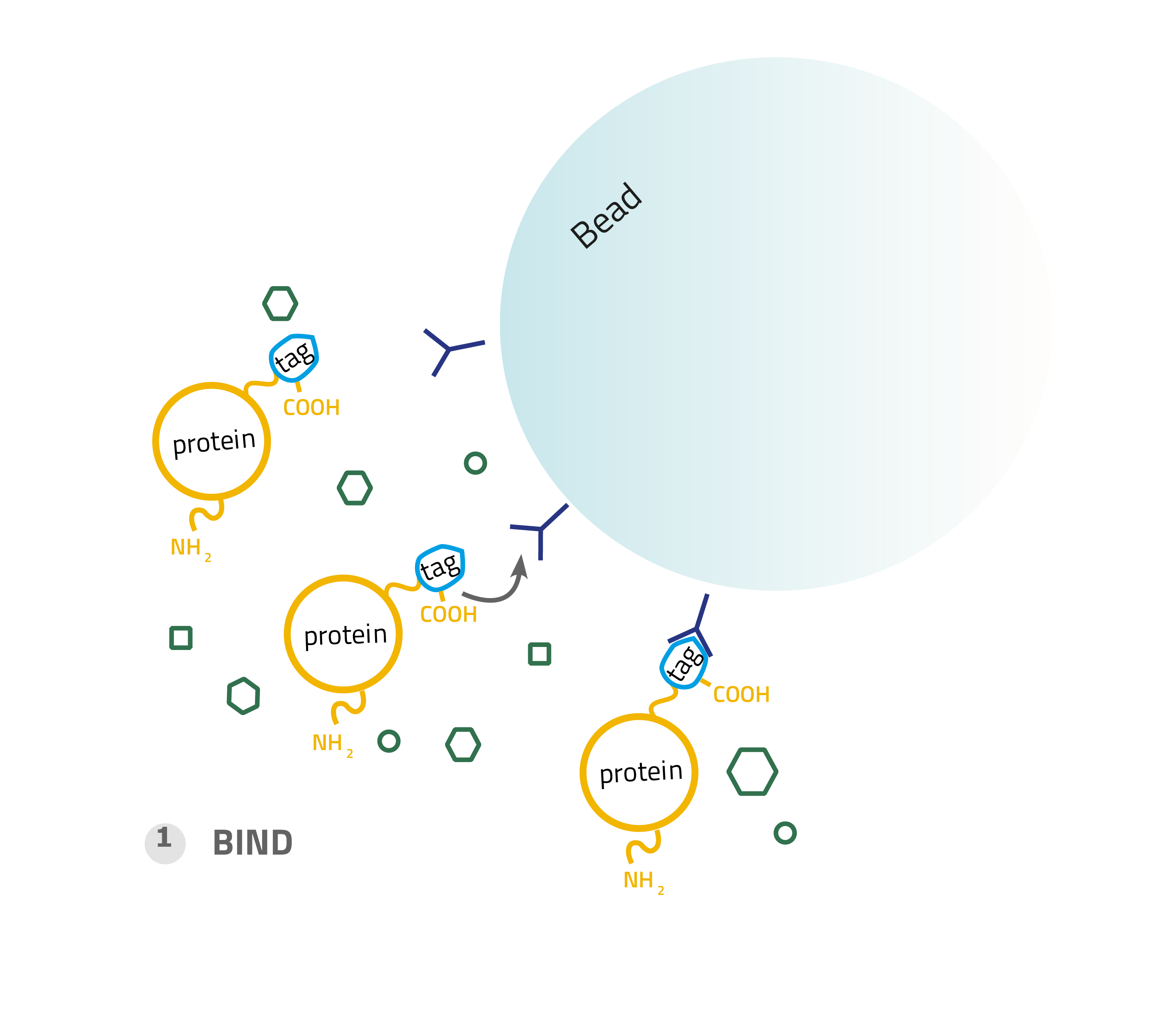
1. Binding:
A complex solution containing the tagged protein is applied to the column and binds based on the affinity tag - matrix interaction
A complex solution containing the tagged protein is applied to the column and binds based on the affinity tag - matrix interaction
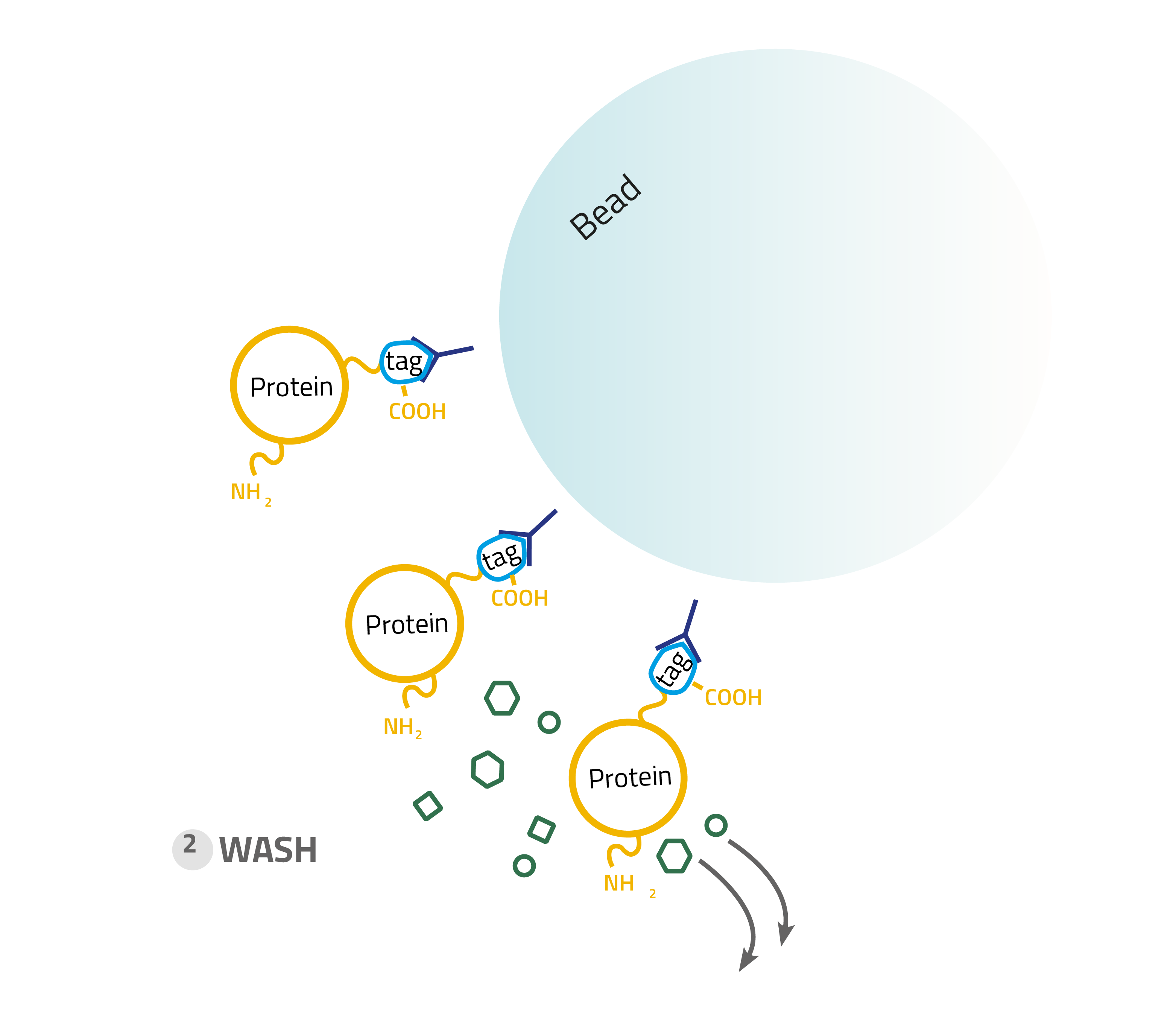
2. Washing:
Other proteins which bind unspecifically are washed away with suitable buffers
Other proteins which bind unspecifically are washed away with suitable buffers
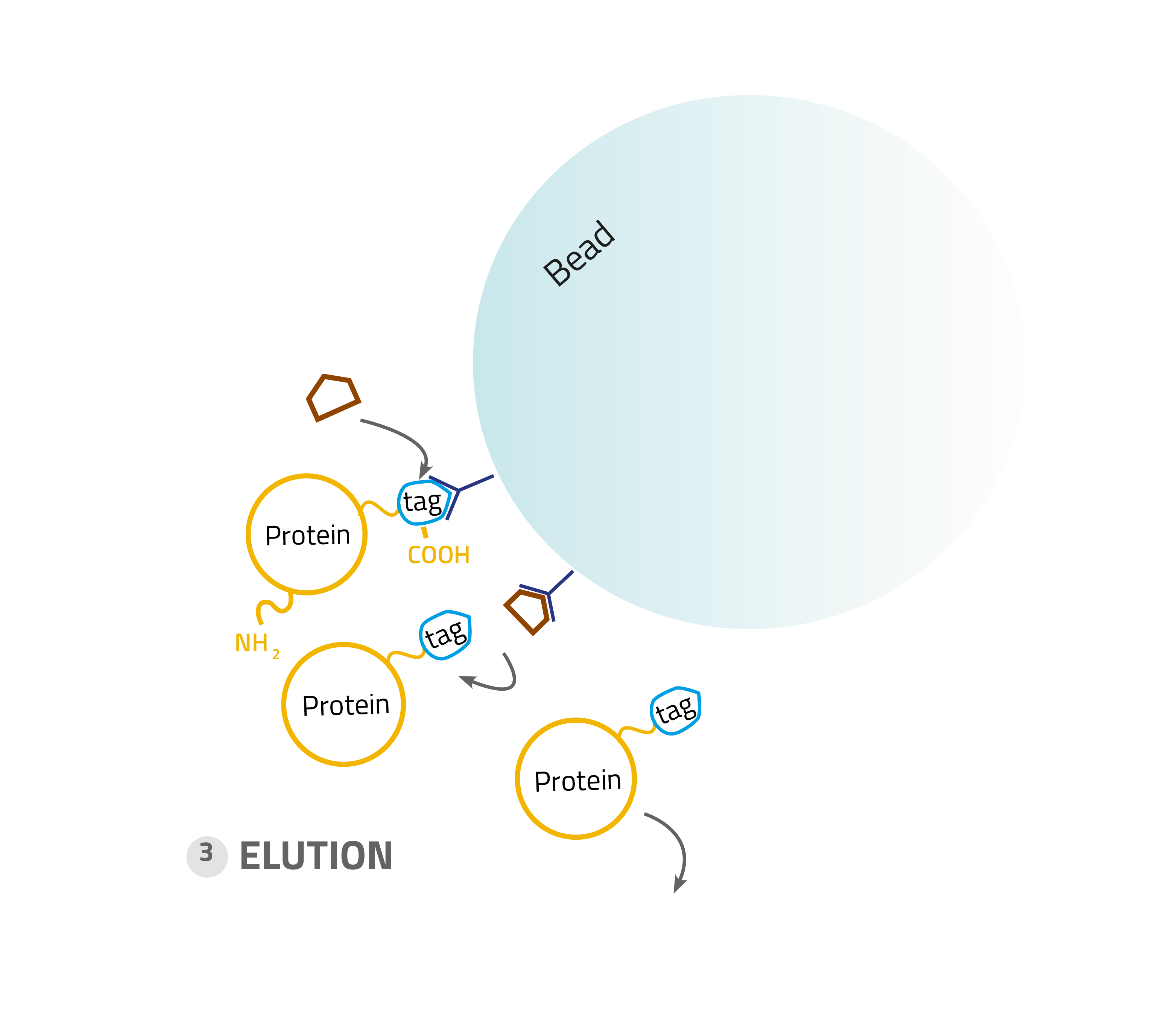
3. Elution:
Specifically bound protein is eluted from the column, typically by competitive binding of a similar molecule (e.g. histidine and imidazole), by cutting off the tag with a protease or by destabilization of the affinity tag - matrix interaction e.g. by a change of pH
Specifically bound protein is eluted from the column, typically by competitive binding of a similar molecule (e.g. histidine and imidazole), by cutting off the tag with a protease or by destabilization of the affinity tag - matrix interaction e.g. by a change of pH
Affinity tag applications
While the term "affinity chromatography" implies that the protein of interest is being purified via the affinity tag, there are a number of applications that can be done in addition to purificaton. These include:
Detection: Specific antibodies are available for most affinity tags, so that tagged proteins can be detected in Western Blots, via immunostaining, in ELISA assays or other antibody-based applications.
Immobilization: Affinity tags can be used to immobilize tagged proteins, e.g. on surface plasmon resonance chips, on ELISA plates or other surfaces. The immobilized proteins can then be assessed e.g. for their ligand binding kinetics.
Pulldown: Affinity-tagged proteins can be pulled down from complex solutions e.g. via affinity magnetic beads. They can also be immobilized via affinity beads and used to pull down interaction partners from complex mixtures, such as cell lysates.
Detection: Specific antibodies are available for most affinity tags, so that tagged proteins can be detected in Western Blots, via immunostaining, in ELISA assays or other antibody-based applications.
Immobilization: Affinity tags can be used to immobilize tagged proteins, e.g. on surface plasmon resonance chips, on ELISA plates or other surfaces. The immobilized proteins can then be assessed e.g. for their ligand binding kinetics.
Pulldown: Affinity-tagged proteins can be pulled down from complex solutions e.g. via affinity magnetic beads. They can also be immobilized via affinity beads and used to pull down interaction partners from complex mixtures, such as cell lysates.
Affinity chromatography methods in the lab
Magnetic Beads
Magnetic Beads Ligands bound to magnetic beads are a fast and smart way to purify proteins (Read the complete guide here). To perform the purification you need a magnetic bead separator which purifies your protein of interest. Our scientist Roland Fabis PhD made a complete MagBead tutorial on YouTube (You can watch it here ).
Magnetic Beads Ligands bound to magnetic beads are a fast and smart way to purify proteins (Read the complete guide here). To perform the purification you need a magnetic bead separator which purifies your protein of interest. Our scientist Roland Fabis PhD made a complete MagBead tutorial on YouTube (You can watch it here ).

FPLC - Fast protein liquid chromatography
FPLC is the standard method for protein purification via affinity chromatography. The advantage of FPLC is that the buffer flow rate is controlled by a positive-displacement pump and the total flow rate of the buffer is kept constant. So FPLC is very suitable for method development and the results are reproducible. The used pressure is with typically less than 5 bar relativley low. The drawback of the method is that you need to buy a very expensive chromatography system.
FPLC is the standard method for protein purification via affinity chromatography. The advantage of FPLC is that the buffer flow rate is controlled by a positive-displacement pump and the total flow rate of the buffer is kept constant. So FPLC is very suitable for method development and the results are reproducible. The used pressure is with typically less than 5 bar relativley low. The drawback of the method is that you need to buy a very expensive chromatography system.
Drip columns
With drip columns you can work directly at your bench. They are small disposable plastic columns packed with chromatography affinity resin. In contrast to the FPLC method you cannot control pressure and flow rate.
With drip columns you can work directly at your bench. They are small disposable plastic columns packed with chromatography affinity resin. In contrast to the FPLC method you cannot control pressure and flow rate.

Batch spin
The batch spin method is done at ambient pressure. The solid phase is packed onto a column and the sample is added to bind the protein. It is a very simple method to get purified proteins in a short time.
The batch spin method is done at ambient pressure. The solid phase is packed onto a column and the sample is added to bind the protein. It is a very simple method to get purified proteins in a short time.
Limitations of affinity chromatography
Since affinity chromatography solely relies on the interaction of a tag-matrix interaction, it is important to be aware of certain limitations:
Tag accessibility
Interaction between the protein tag and the purification matrix is only possible if the tag is accessible and not buried within the folds of the target protein. Other buffer components such as detergents can also bury the tag sequence and have a negative effect on the tag/matrix interaction. In many cases, moving the tag to the other terminus of the protein - or even to an accessible loop within the protein - can help. If this proves unsuccessful, protein purification via the His-tag (but not via other tags) can also be done under denaturing conditions, thereby exposing the tag to the matrix surface.
Influence of tag on expression
Depending on the protein, tags may interfere with expression rates, folding, and activity. This effect is rather inpredictable unless structural information is already available. Therefore, screening of different tags and tag positions can be useful.
Discrimination between protein conformations / folding variants
Affinity matrices purify all proteins which carry the affinity tag, regardless of correct folding or conformation. To discriminate between different protein conformations, multimers, or to separate correctly folded from inactive protein, protein-specific purification matrices are a useful alternative or additional method.
Tag accessibility
Interaction between the protein tag and the purification matrix is only possible if the tag is accessible and not buried within the folds of the target protein. Other buffer components such as detergents can also bury the tag sequence and have a negative effect on the tag/matrix interaction. In many cases, moving the tag to the other terminus of the protein - or even to an accessible loop within the protein - can help. If this proves unsuccessful, protein purification via the His-tag (but not via other tags) can also be done under denaturing conditions, thereby exposing the tag to the matrix surface.
Influence of tag on expression
Depending on the protein, tags may interfere with expression rates, folding, and activity. This effect is rather inpredictable unless structural information is already available. Therefore, screening of different tags and tag positions can be useful.
Discrimination between protein conformations / folding variants
Affinity matrices purify all proteins which carry the affinity tag, regardless of correct folding or conformation. To discriminate between different protein conformations, multimers, or to separate correctly folded from inactive protein, protein-specific purification matrices are a useful alternative or additional method.
Further reading
- IDA vs NTA: A tale of two ligands, Cube Biotech 2013
- Angelo DePalma, PhD: Poly-His Tags Improve Protein Purification. The most popular protein tagging method has many advantages and few caveats. Genetic Engineering and Biotechnology News May 8, 2014
- Angelo DePalma, PhD: Getting a Sure Hold on Protein Purification by Attaching Convenient Handles. Genetic Engineering and Biotechnology News Mar 1, 2015, Vol. 35, No. 5
- Locatelli-Hoops, S.C. et.al. 2013. Expression, surface immobilization, and characterization of functional recombinant cannabinoid receptor CB2. Biochim. Biophys. Acta 1834 (10):2045-56.
- PureCube Rho1D4 Agarose, Cube Biotech 2013 Trademarks and disclaimers: StrepTactin® and Strep-tag®(IBA), FLAG® (Sigma-Aldrich Co. LLC) membranes11060451


